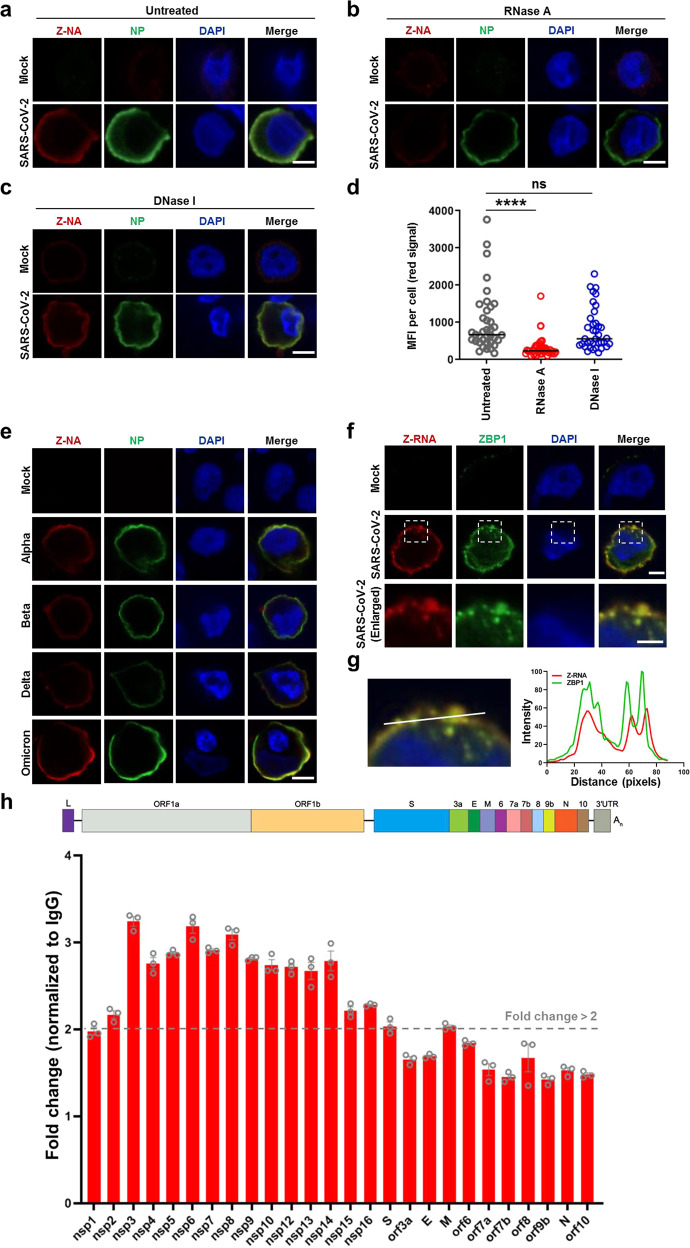
Fig. 2. Formation of Z-RNA during SARS-CoV-2 infection.
a–d SARS-CoV-2 original strain (MOI = 0.1) infected or uninfected (mock) Calu-3 cells were fixed and permeabilized at 48 hpi, and then either untreated (a) or treated with RNase A (b) or DNase I (c). Cells were co-stained with antibodies against Z-NA (red) and viral antigen NP (green). Nuclei were stained by DAPI (blue). d Quantification of the median florescence intensity (MFI) of Z-NA staining in SARS-CoV-2-infected cells in a–c. Comparison of median values between two groups was analyzed by Mann–Whitney test. ****P < 0.0001; ns, no significance. Scale bar, 10 μm. e Calu-3 cells were infected with SARS-CoV-2 Alpha, Beta, Delta and Omicron strains (MOI = 0.1) for 48 h, and stained with antibodies against Z-NA (red) and NP (green). Scale bar, 10 μm. f SARS-CoV-2 original strain infected Calu-3 cells were co-stained with Z-NA and ZBP1 antibodies. Scale bars, 10 μm and 5 μm in enlarged image. g Quantitative analysis of the co-localization between Z-RNA and ZBP1 was performed with ImageJ. h Identification of viral derived Z-RNA. Calu-3 cells infected with SARS-CoV-2 (MOI = 0.1) for 48 h were lysed and incubated with Z-RNA antibody or isotype mouse IgG for immunoprecipitation. Enriched RNA was subjected to qRT-PCR analysis.