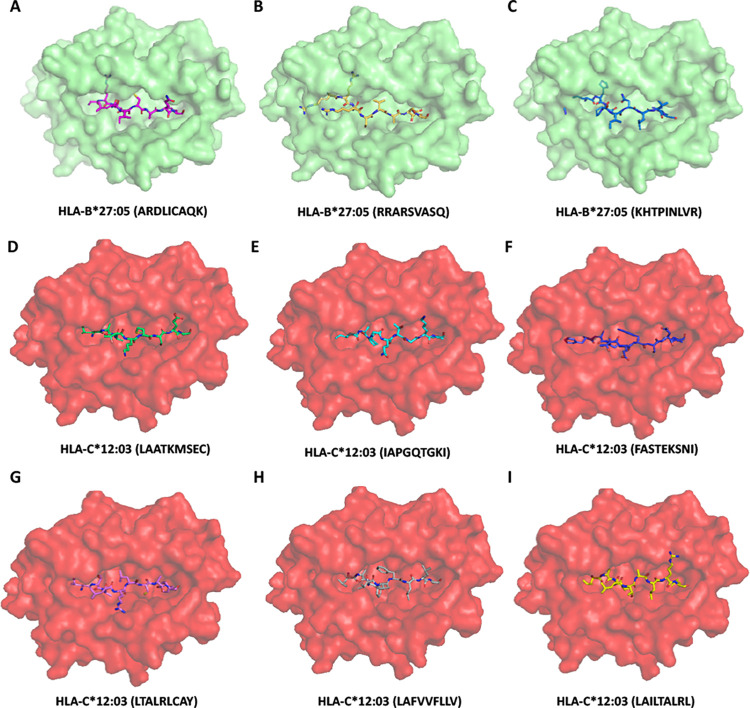
Fig 4. Model of class I HLA molecule associated with protective (HLA-B*27:05) or risk (HLA-C*12:03) incidence of SARS-CoV-2 severity.
(A-C) HLA-B*27:05 binds to ARDLICAQK, RRARSVASQ and KHTPINLVR derived from the S protein with affinity estimated Kd 1534 nM, 438.61 nM, 6798.81 nM respectively. (D-F) HLA-C*12:03 binds to LAATKMSEC, IAPGQTGKI and FASTEKSNI derived from the S protein with affinity estimated Kd 959.47 nM, 806.95 nM, 126.76 nM respectively. (G-I) HLA-C*12:03 binds to LTALRLCAY, LAFVVFLLV and LAILTALRL derived from the E protein with affinity estimated Kd 176.42 nM, 190.66 nM, 112.95 nM respectively. Peptides are shown as sticks modeled on the crystal structure of class I HLA molecues, different peptide backbones are represented by different colors.