Figure 2. Benchmarking performance across a range of study designs.
Values represent the average of three independent trials. FDR: False Discovery Rate; TPR: True Positive Rate. For phasing and imputation, gray indicates that no hetSNPs remained after downsampling. For meiotic recombination discovery, gray indicates the absence of a prediction class (e.g., zero FNs, FPs, TNs, or TPs). Simulations roughly matching the characteristics of the Sperm-seq data are outlined in red.
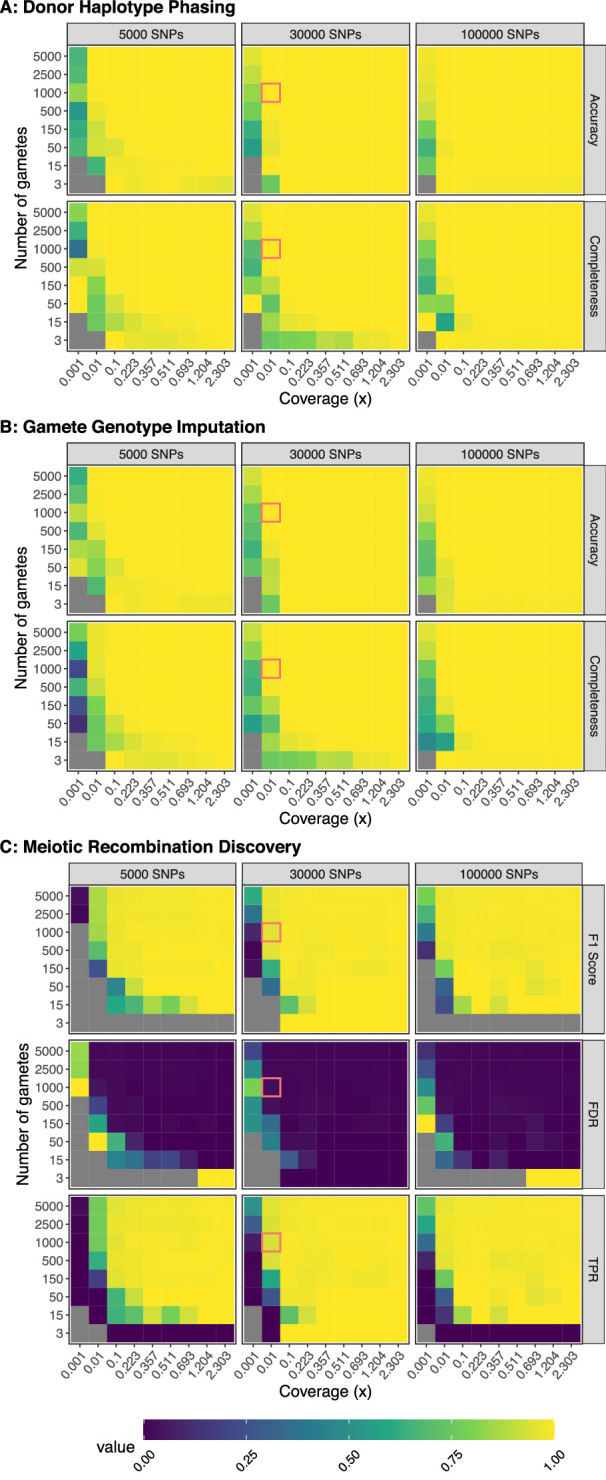
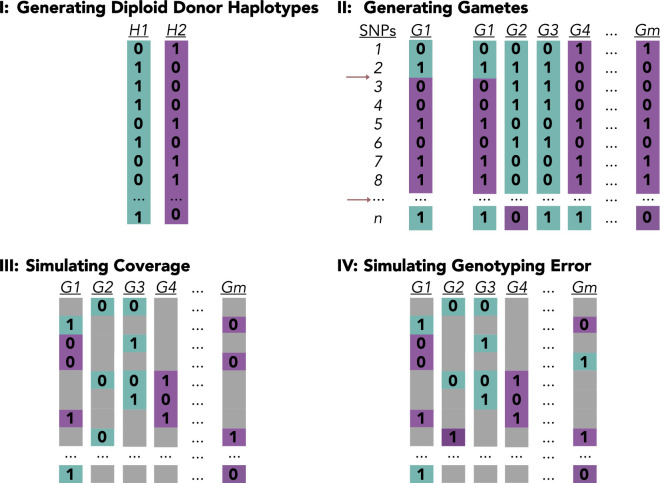
Figure 2—figure supplement 1. Generative model.
Figure 2—figure supplement 2. Benchmarking performance across a range of study designs - Additional Metrics.
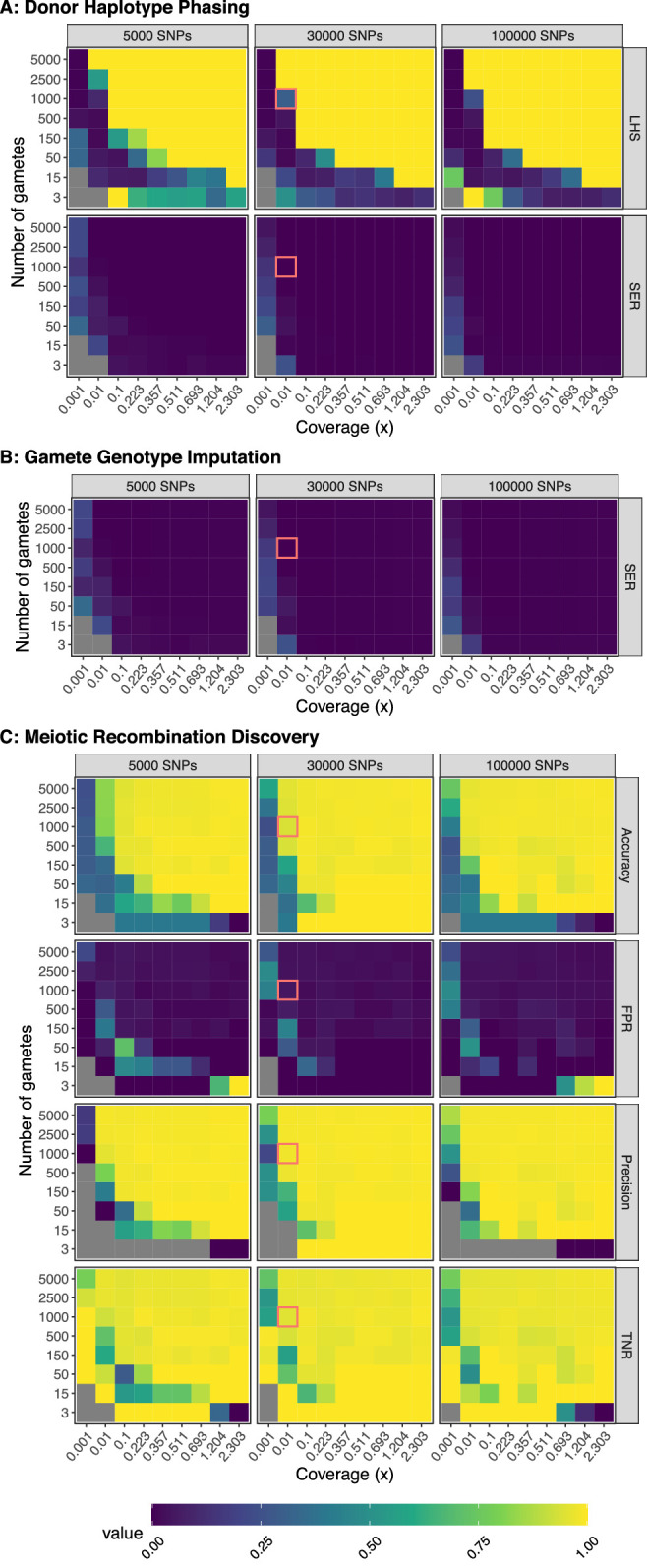
Figure 2—figure supplement 3. Illustration of switch error rate (SER) and accuracy.

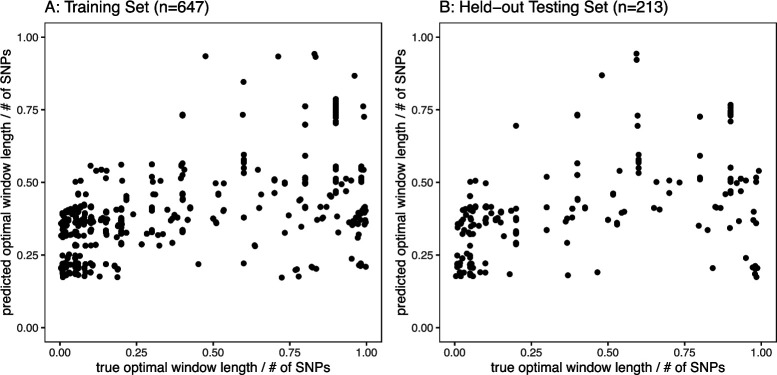
Figure 2—figure supplement 4. Automatic phasing window size calculation.
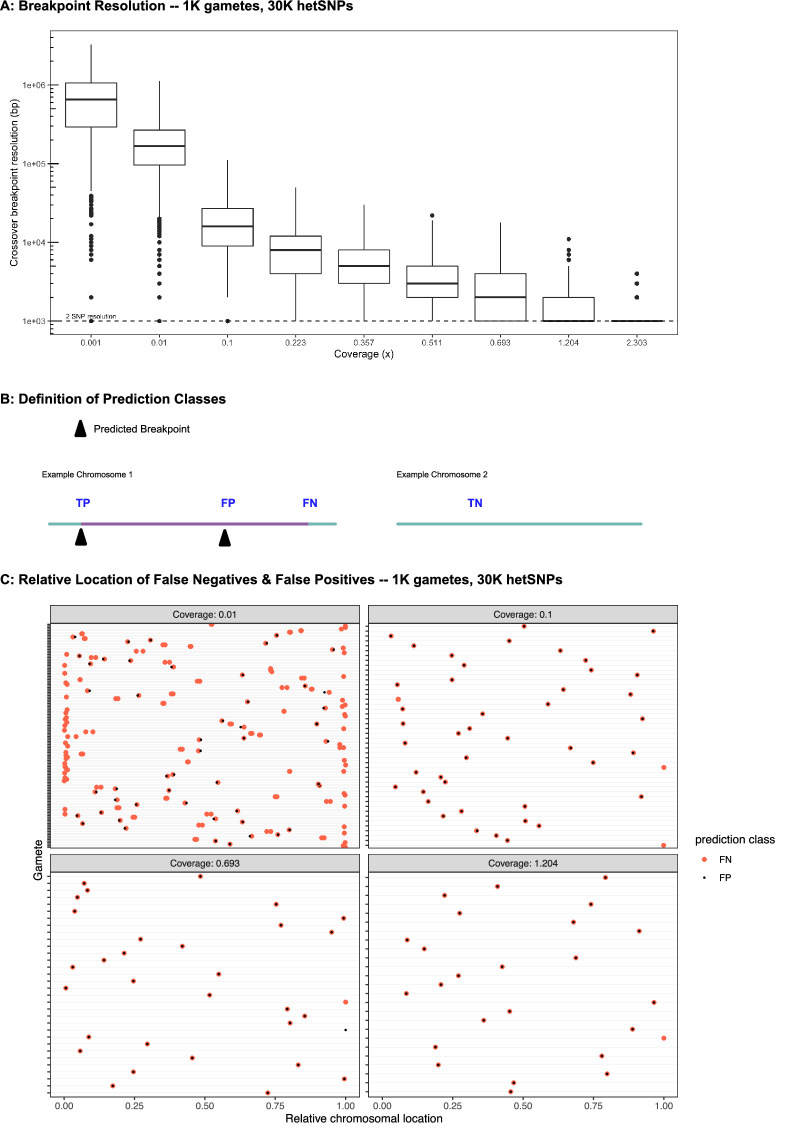
Figure 2—figure supplement 5. Discovery of meiotic recombination events in simulated data reflecting characteristics of the Sperm-seq data.
Figure 2—figure supplement 6. Model robustness when genotyping error is underestimated.
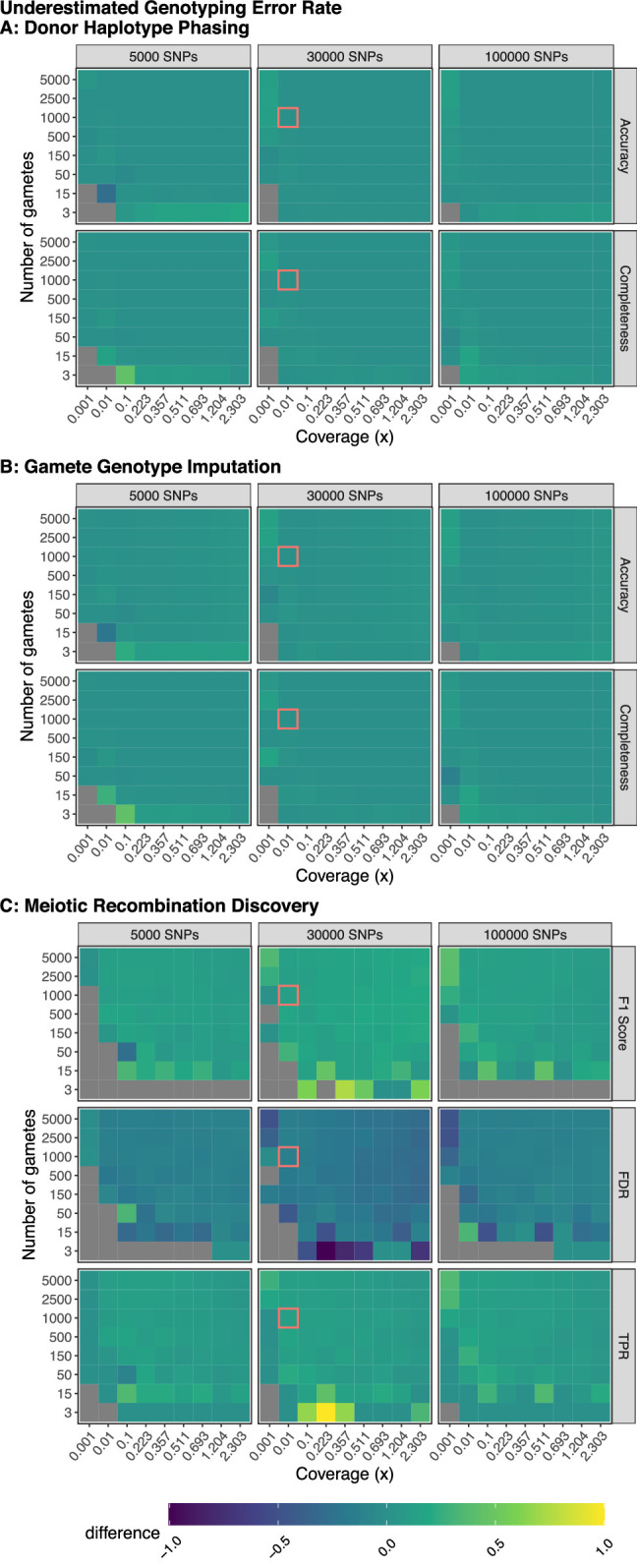
Figure 2—figure supplement 7. Model robustness when recombination rate is underestimated.
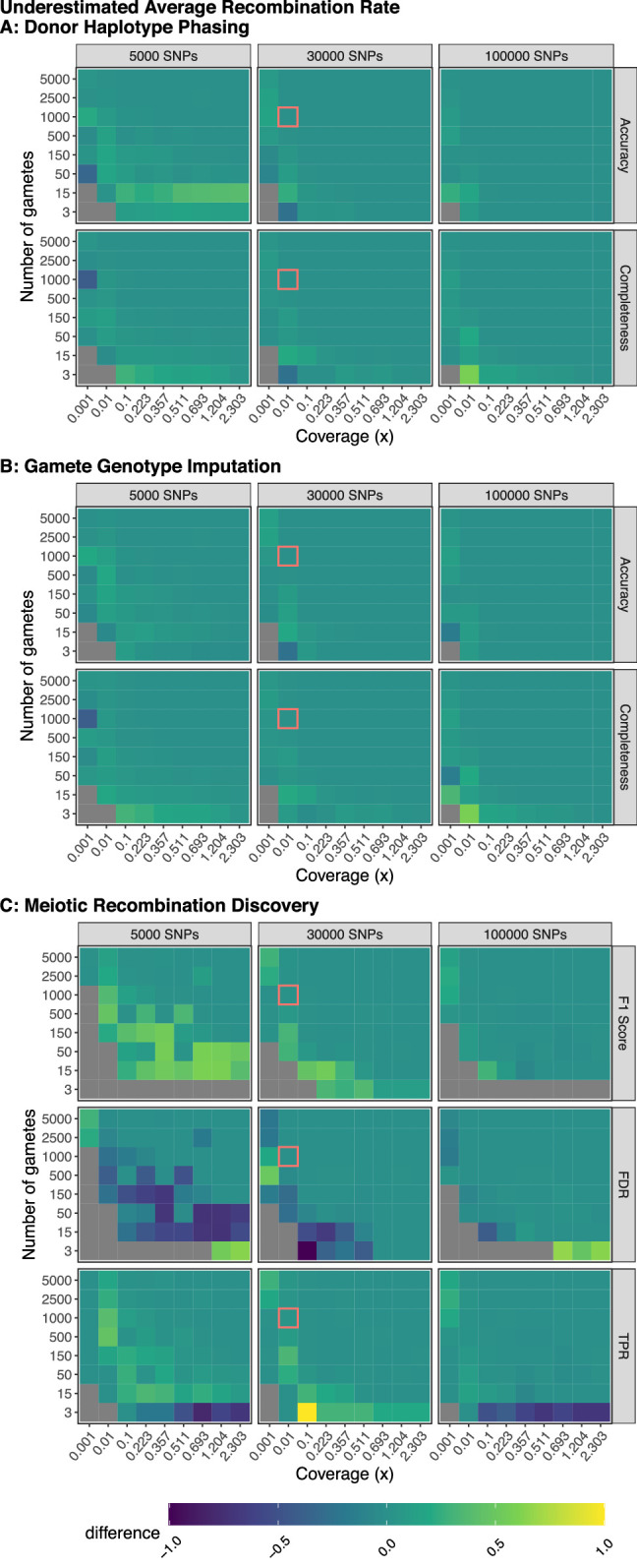
Figure 2—figure supplement 8. Model robustness when genotyping error is overestimated.
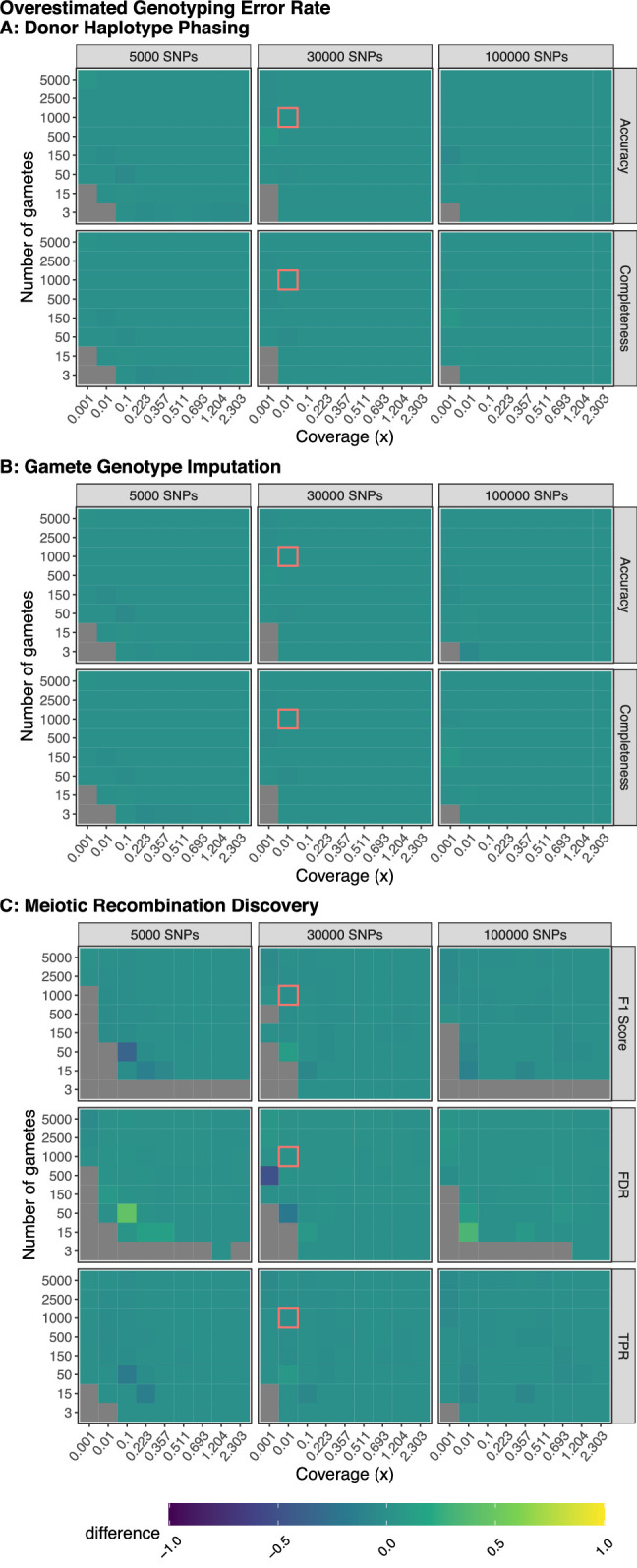
Figure 2—figure supplement 9. Model robustness when recombination rate is overestimated.
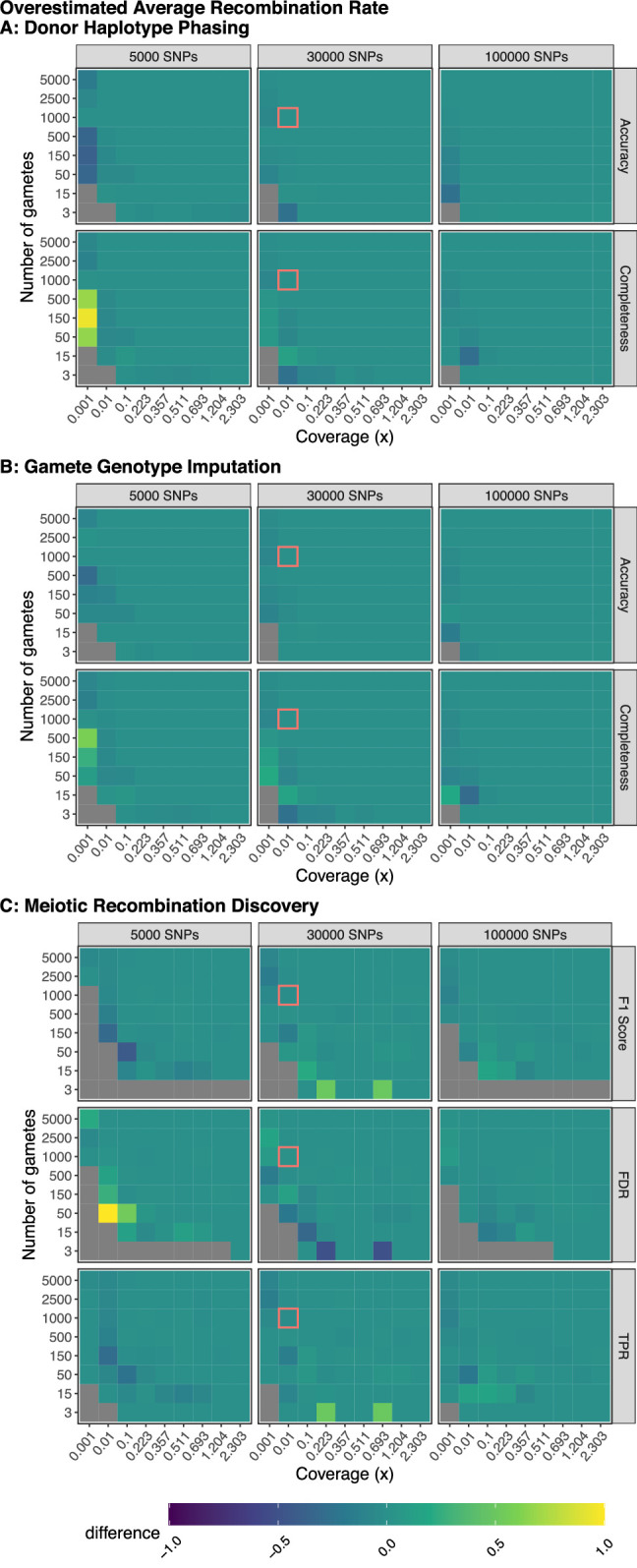
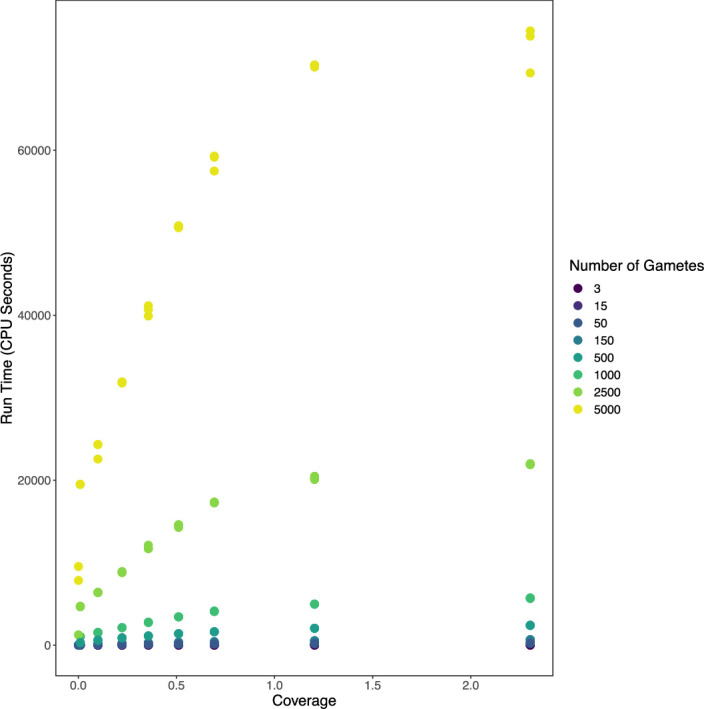
Figure 2—figure supplement 10. Benchmarking rhapsodi runtime across a range of simulated data profiles.