Figure 2. Heterologous expression of modified PedA15 peptides in E. coli and analysis by MALDI-TOF MS.
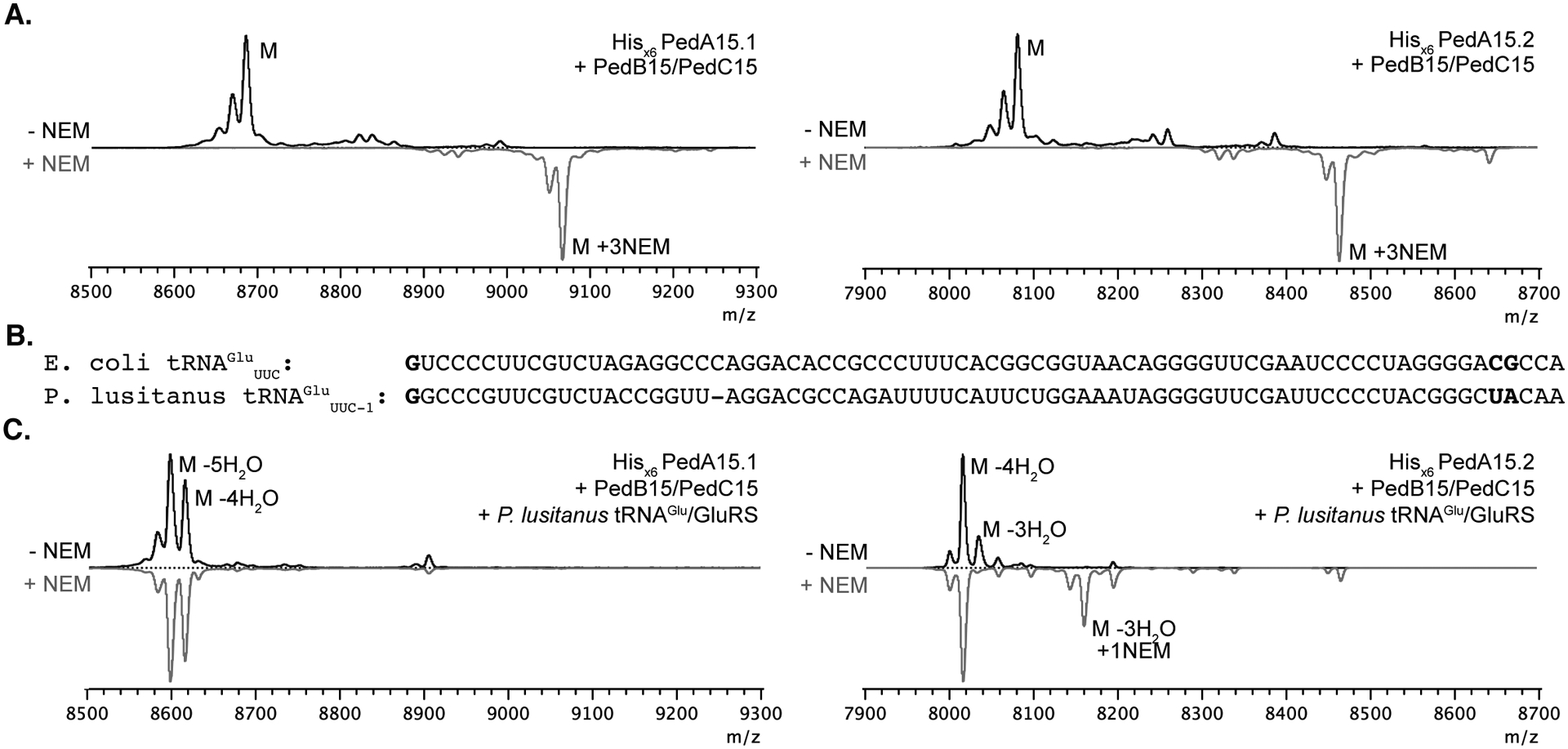
(A) Coexpression in E. coli BL21 (DE3) of PedA15 precursor peptides, PedB15 and PedC15 in the absence of P. lusitanus tRNAGlu/GluRS. Both PedA15.1 (left; avg. m/z 8687.7 calc.; 8686.6 obs.) and PedA15.2 (right; avg. m/z 8085.0 calc.; 8085.4 obs.) were unmodified in the absence of aminoacyl-tRNA from the native producer. Consequently, NEM treatment of these samples resulted in full alkylation of all Cys residues (grey spectra). (B) Comparison of tRNAGlu sequences in E. coli (Proteobacterium) and P. lusitanus NL19 (Bacteroidetes). The anticodon is denoted in subscript. Nucleotide positions previously shown to be important for LanB compatibility are in bold.15 (C) PedA15 precursor peptides co-expressed with PedB15, PedC15 and P. lusitanus tRNAGlu/GluRS. Isolated product for PedA15.1 (left) contained a mixture of 4x (avg. m/z 8615.7 calc.; 8615.8 obs.) and 5x (avg. m/z 8597.6 calc.; 8597.0 obs.) dehydrated peptide and was fully cyclized as evidenced by a lack of NEM alkylation. PedA15.2 (right) was purified as a mixture of 3x (avg. m/z 8030.9 calc.; 8030.5 obs.) and 4x dehydrated (avg. m/z 8012.9 calc.; 8012.7 obs.) peptide, with a minor amount of partially cyclized peptide product observed by NEM alkylation. M, parental mass (avg. m/z, unmodified peptide); NEM, N-ethylmaleimide (grey traces); “-n” H2O, indicates the number (n) of dehydrations consistent with the m/z of the labeled peak.
