Figure 4. NMR analysis of cyclized PedA15.1 core peptide.
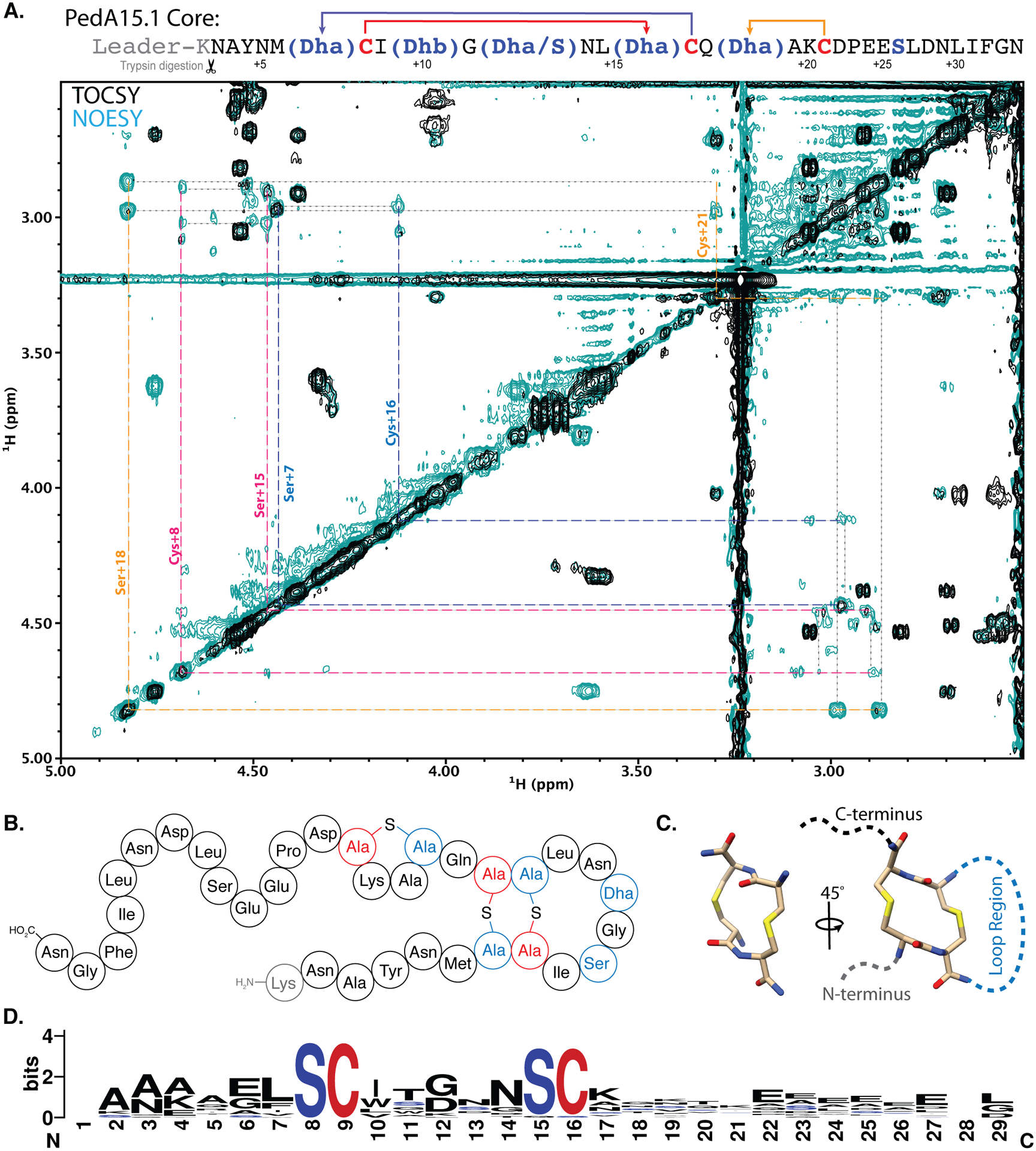
(A) 2D-NMR overlay (TOCSY, black; NOESY, teal) of the aliphatic region highlighting CH2-S-CH2 intra-bridge correlations used to infer lanthionine ring pattern. Intra-bridge NOEs are denoted with black dotted lines. Residues and lanthionine ring systems are color coded according to the sequence diagram at the top. The NMR data also showed that the difference between 4 and 5-fold dehydrated peptide is at Ser12, and that this residue is not involved in ring formation. For full structural assignment data, see Supporting Information. (B) Diagram representing the pattern of cyclized PedA15.1 that is dehydrated five times, as determined by NMR and MS/MS analysis (C) Representation of the unusual 14-membered bis-lanthionine heterocycle identified in the PedA15 peptides. 3D model generated in Avogadro86 representing the LL-Lan/LL-Lan stereochemical configuration. (D) Sequence logo for the core peptide of a family of class I lanthipeptide precursor peptides encoded in Bacteroidetes. See Figure S13 for individual peptides. The SCXnSC motif (n = 3–6, Fig. S13) is highlighted in red and blue.
