FIGURE 5.
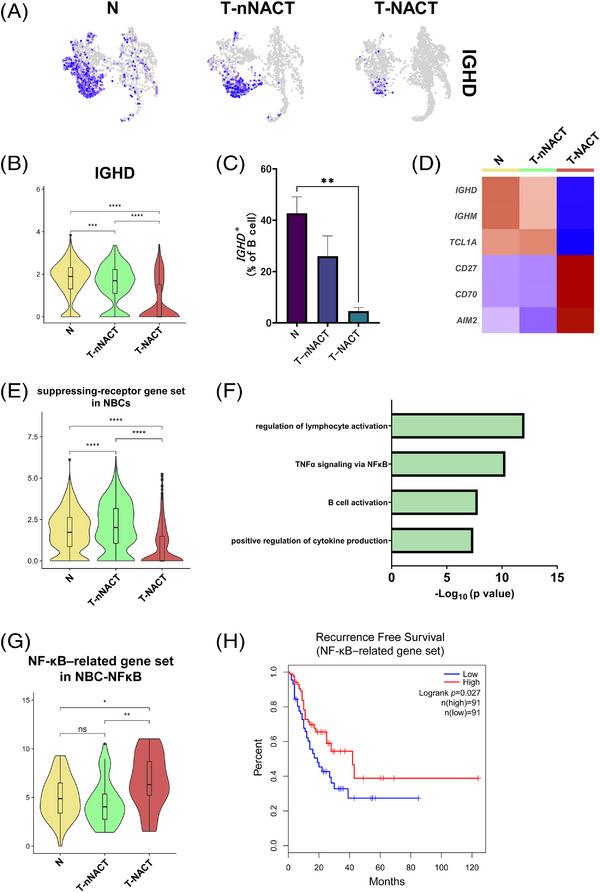
Decrease of the proportion and functional changes in NBCs during chemotherapy. (A) UMAP plots showed expression levels of IGHD, which is a canonical marker gene for naive B cells (NBCs), by the tissue source type. (B) Violin plots showed the expression of IGHD in NBCs by the tissue source type. Significance of IGHD expression (p value) between tissue source types was determined by Wilcoxon rank‐sum test. Boxplots included centreline, median; box limits, upper and lower quartiles; whiskers at most 1.5× the interquartile range past upper and lower quartiles. *** p < .001, **** p < .0001. (C) Bar plots showed the percentage of IGHD + cells to total B cells by the tissue source type (N; n = 7, T‐nNACT; n = 5, T‐NACT; n = 5). Mean + SEM shown. Dunn's multiple comparisons test was performed. ** p < .01. (D) Heatmap showed the expression of DEGs among tissue source types in NBCs. (E) Violin plots showed the expression of the suppressing‐receptor gene set in NBCs. Significance of this gene set enrichment (p value) between tissue source types was determined by Wilcoxon rank‐sum test. Boxplots included centreline, median; box limits, upper and lower quartiles; whiskers at most 1.5× the interquartile range past upper and lower quartiles. **** p < .0001. (F) The enriched Gene Ontology term for the top 100 signature genes in the NBC subtype with T‐NACT using Metascape. (G) Violin plots showed the expressions of the NF‐κB‐related gene set by the tissue source type. Significance of this gene set enrichment (p value) between tissue source types was determined by Wilcoxon rank‐sum test. Boxplots included centreline, median; box limits, upper and lower quartiles; whiskers at most 1.5× the interquartile range past upper and lower quartiles. ns; not significant, * p < .05, ** p < .01. (H) Survival analysis based on the NF‐κB‐related gene set in TCGA‐ESCA (n = 182). Log‐rank Mantel Cox–test.
