Figure 2. Establishing titratable CRISPRi with mismatch sgRNA libraries.
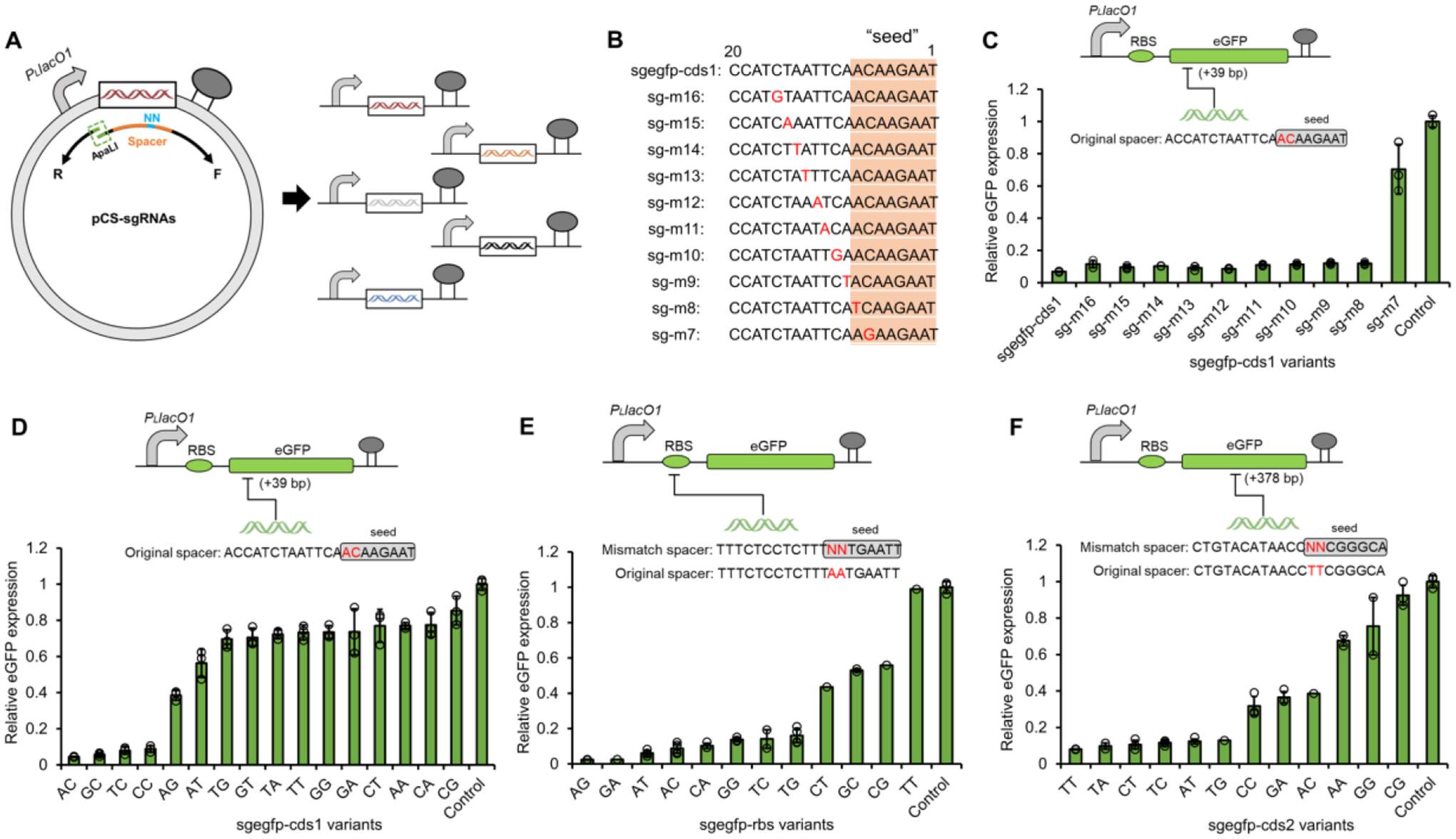
(A) The design of inverse PCR for one-pot construction of mismatch sgRNAs. (B) Spacer sequences (5’-3’) of single mismatch sgegfp-cds1 variants targeting the coding sequence (+39 bp) of eGFP. (C) The CRISPRi efficiencies of sgRNAs with single mismatch at different positions in the sgegfp-cds1 spacer (c), with rationally designed double mismatches in the 7–8th bp seed region of sgegfp-cds1 spacer (D), with randomly mutated double mismatches in the 7–8th bp seed region of sgegfp-rbs spacer (targeting the RBS region eGFP promoter) (E), and with randomly mutated double mismatches in the 7–8th bp seed region of sgegfp-cds2 spacer (targeting the coding sequence (+378 bp) of eGFP) (F). Control, E. coli∷dCas9 co-transformed with pZE-eGFP and pCS27 empty plasmid. In E and F, 32 single colonies were randomly picked from each plate and were subjected to both eGFP fluorescence assay and spacer determination via plasmid sequencing. All data in C and D represent the mean of three biologically independent samples and error bars show standard deviation.
