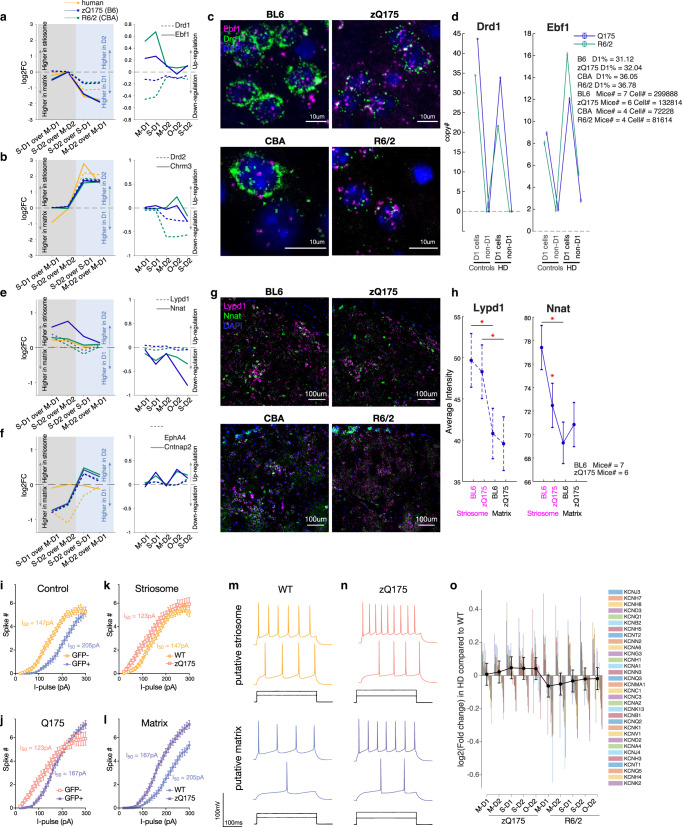
Fig. 6. Histological and physiological loss of compartmental identity in HD model mice.
a snRNA-seq data for dSPN markers, i.e., Drd1 (broken lines) and Ebf1 (solid lines). Left: Differential expression was measured between the cell-type pairs indicated below and shown as log2(fold change). Right: Expressions in HD models are compared to those in controls. b Same as in a, but for iSPN markers, i.e., Drd2 (broken lines) and Chrm3 (solid lines). c FISH images of sections obtained from the anterior striatum in the two HD models compared to their controls, stained for Drd1 (green), Ebf1 (magenta), and DAPI (blue). d Quantification of FISH image. Copy numbers (i.e., number of detected spots) for Drd1 (left) and Ebf1 (right) are shown separately for D1 and non-D1 cells in controls or HD models. Error bars indicate 95% confidence intervals from the averages. e, f Same as in a and b, but for striosome markers (e), i.e., Lypd1 (broken lines) and Nnat (solid lines), or matrix markers (f), i.e., EphA4 (broken lines) and Cntnap2 (solid lines). g Same as in c but for Lypd1 (magenta), Nnat (green), and DAPI (blue). The images were obtained from anterior striatal sections of the same mice shown in c. h Quantification of FISH image. Average intensity of FISH signals is shown for Lypd1 (left) and Nnat (right) separately for striosomes and matrix in zQ175 mice and in their controls. Error bars indicate 95% confidence intervals. One-way ANOVA followed by Tukey-Kramer post-hoc multiple comparison test. One-way ANOVA: p = 0.0001, multiple comparison: striosomes vs. matrix in BL6; p = 0.0017, striosomes vs. matrix in zQ175; p = 0.0073 for Lypd1 (left). One-way ANOVA: p = 5.89 × 107, multiple comparison: BL6 vs. zQ175 in striosomes, p = 0.0052 for Nnat (right). i–l Using zQ175 mice crossed with a matrix reporter mouse line (CalDAG-GEFI-GFP), electrophysiological properties of putative striosomal (GFP-negative, orange) and putative matrix (GFP-positive, purple) SPNs were examined ex vivo. Current-frequency responses are shown for control (i) and zQ175 (j) mice, or for putative striosomal SPNs (k) and putative matrix SPNs (l). I50 is defined as the input current producing 50% of maximal spike numbers. Error bars indicate SEM. Control: N = 10 mice, n = 212 neurons. zQ175: N = 9 mice, n = 198 neurons. The mean ± SD number of cells recorded per each mouse evaluated was 22 ± 6. m Representative traces in control mice are shown separately for putative striosomal (top, orange) and putative matrix (bottom, purple) SPNs in response to I50 and I90 of putative striosomal SPNs. n Same as in m, but for zQ175 mice. o Dysregulation of potassium channels separately shown for each cell-type of R6/2 and zQ175 mice as compared to controls. We included all 32 out of 79 potassium channels whose dysregulations were quantifiable. Error bars indicate 95% confidence intervals from the averages. See also Supplementary Fig. 8.