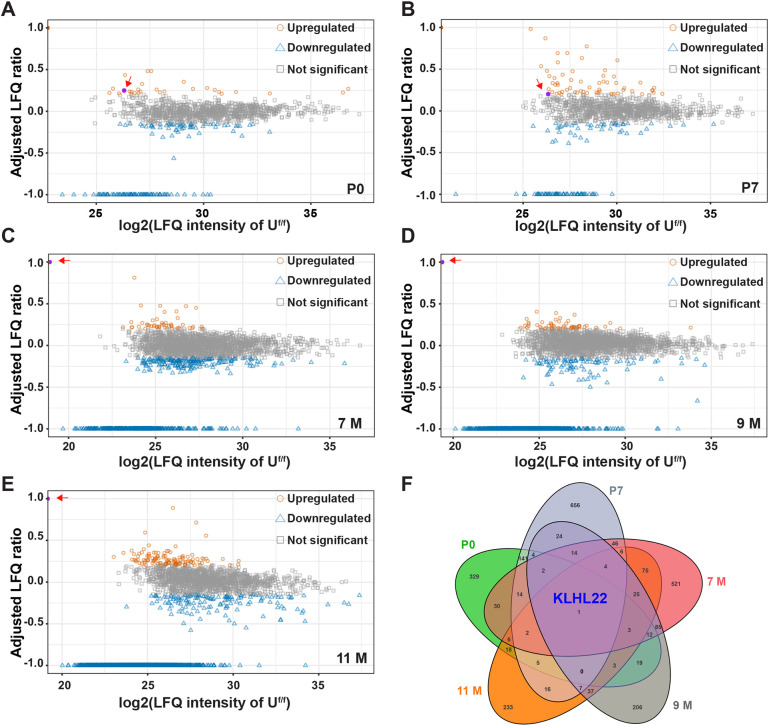
Fig. 2.
Deletion of UBE4B causes upregulation of KLHL22 in neonatal and mature brains as revealed by proteomic analyses. (A,B) Mass spectrometry analysis of postnatal whole brain proteomes following embryonic deletion of UBE4B in NSCs. The x-axis of the scatter plots represents log2-transformed label-free quantification (LFQ) intensities of each protein in Ube4bf/f. The y-axis represents adjusted ratio of LFQ intensities between Ube4bf/f; Nestin-Cre/+ versus Ube4bf/f. Three pairs of littermates at P0 (A) and P7 (B) were analyzed, respectively. Commonly upregulated proteins in Nestin-CKO brains from both P0 and P7 are represented by orange circles and commonly downregulated genes are represented by blue triangles. Red arrows point to KLHL22, represented by a purple circle. Details of data analyses are described in Materials and Methods. (C-E) Mass spectrometry analysis of forebrain proteomes from mature adults following postnatal deletion of UBE4B. The x-axis of the scatter plots represents log2-transformed LFQ intensities of each protein in Ube4bf/f. The y-axis represents adjusted ratio of LFQ intensities between Ube4bf/f; Camk2a-Cre/+ and Ube4bf/f. Three pairs of littermates at 7 months (C), 9 months (D) and 11 months (E) of age were analyzed, respectively. Commonly upregulated proteins in Camk2a-CKO animals from all three age groups are represented by orange circles and commonly downregulated genes are represented by blue triangles. Red arrows point to KLHL22, represented by a purple circle. (F) Venn diagram showing that KLHL22 is the sole protein upregulated upon UBE4B knockout in all five age groups.