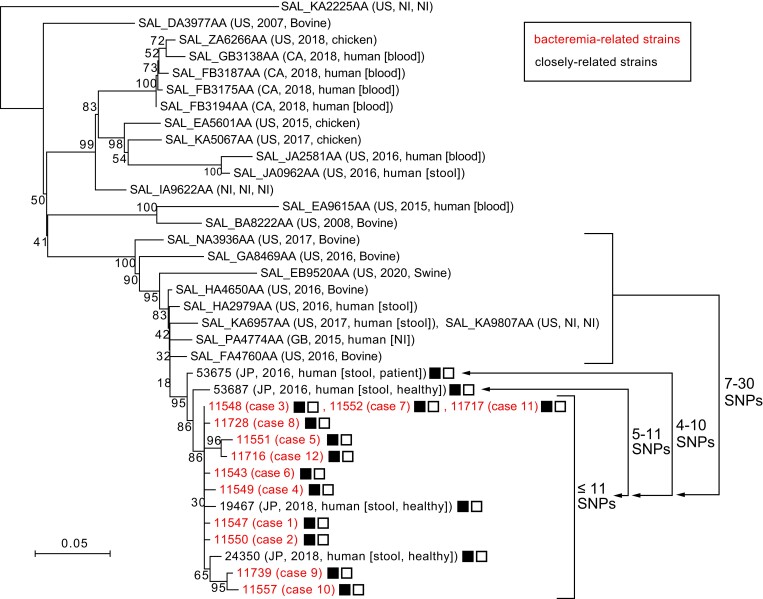
Figure 3.
Core genome–based phylogenetic analysis of the 12 bacteremia-related strains and the 26 closely related strains. Based on the 276 single-nucleotide polymorphisms (SNPs) identified in the core genome (4 686 981 bp in total length), a neighbor-joining tree was constructed based on the identified core genome SNPs using the p-distance method in MEGA7. Strain names (Japanese strains) or strain IDs in the EnteroBase database are indicated along with other strain information (country, isolation year, and source). The maximum SNP distance within the subcluster that included bacteremia-related strains was 11. The ranges of SNP distances from the subcluster to the 2 most closely related strains are also indicated. The presence of the plasmids pSO11548-1 (3428 bp in size) and pSO11548-2 (2495 bp in size) is indicated by solid and outlined squares, respectively. Abbreviation: NI, no information.