Figure 10.
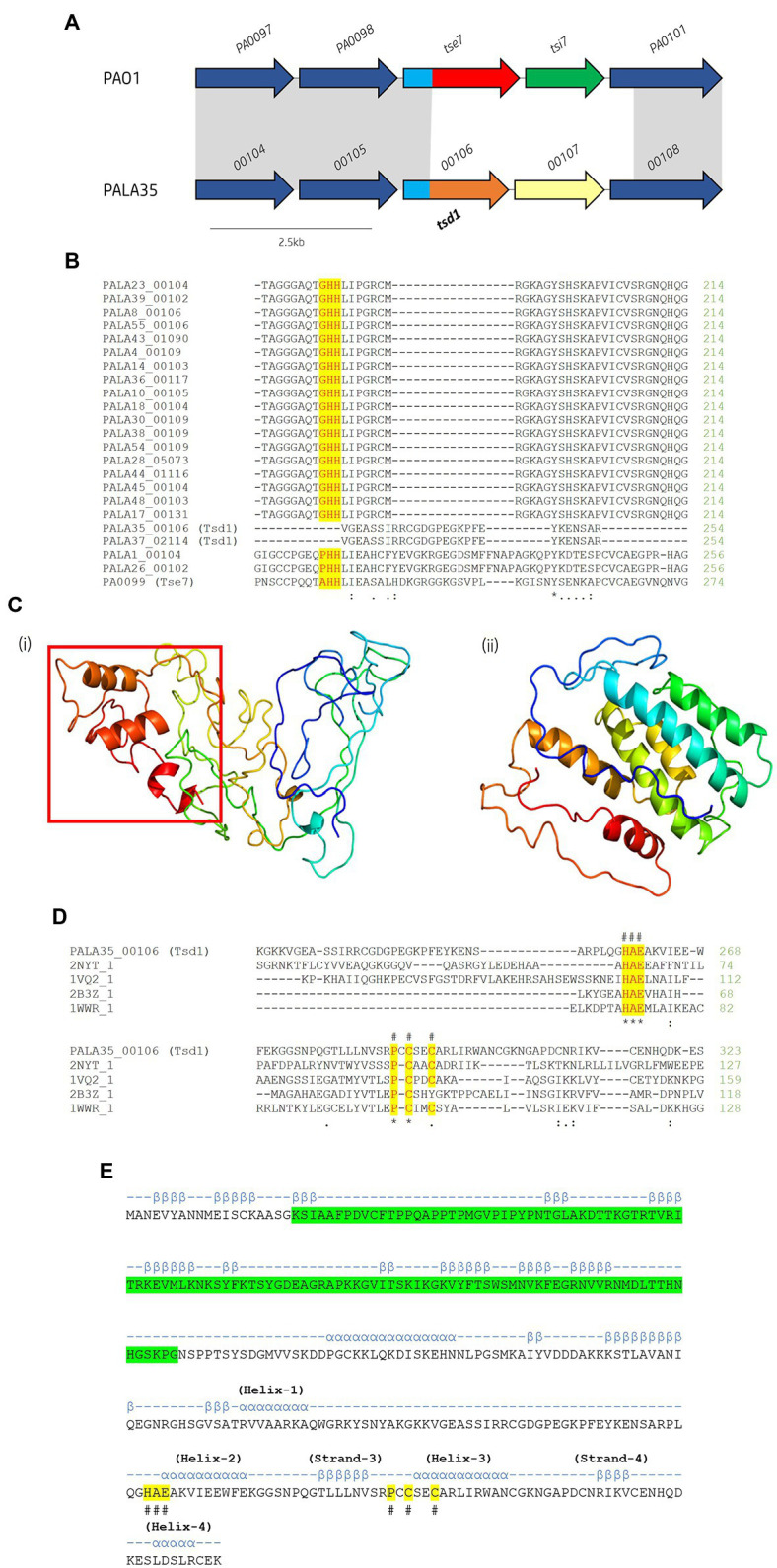
In silico analysis of putative cytidine deaminase Tsd1. (A) Schematic of the orphan effector-immunity island tse7-tsi7 in reference P. aeruginosa and representative clinical isolate PALA35. Homologous genes are connected by grey bands. Proteins of unknown function are coloured dark blue. The N-terminal PAAR-like domain DUF4150 is light blue [aa 1-135]. The variable C-terminal is coloured red for the domain Tox-GHH2 in PA0099/tse7 [aa 135-315] and light brown for the C-terminal cytidine deaminase domain in PALA35_0106/tsd1 [aa 135-331]. Cognate immunities are coloured green for PA0100/tsi7 and yellow for PALA35_0107/tsdI1. (B) Partial amino acid sequence alignment of Tse7 homologs from the clinical isolates to Tse7 (PA0099) from reference strain PAO1. Residues of the characteristic GHH motif of Tox-GHH2 are highlighted yellow. Note the absence of the GHH motif from PALA35/36 Tsd1 suggesting Tsd1 has a different mechanism of action (C) Phyre2 predicted models of (i) Tsd1 and (Continued)FIGURE 10 (Continued)(ii) TsdI1. The red box highlights the location of the four α helical toxic activity-containing C-terminus. (D) Tsd1 is a putative deaminase as shown by amino acid alignment of Tsd1 from isolate PALA35 (00106) to deaminase APOBEC-2 from Homo sapiens (PDB: 2NYT_1), T4-bacteriophage deoxycytidylate deaminase from Escherichia virus T4 (PDB: 1VQ2_1), bifunctional deaminase Riboflavin biosynthesis protein RibD from Bacillus subtilis (PDB: 2B3Z_1), and tRNA adenosine deaminase TadA from Aquifex aeolicus (PDB: 1WWR_1). Residues of the HAE and PCxxC deaminase motifs are highlighted yellow and indicated by an #. Additional symbols are the same as in Figure 8. (E) Tsd1 encodes a putative PAAR-like DUF4150 domain in addition to the deaminase motifs. PSIPred secondary structure prediction of Tsd1 with the PAAR-like domain DUF4150 (PF13665) highlighted in green, alpha-helices are annotated by α and Beta-strands by β. The location of important α and β structure of the cytidine deaminases superfamily as described by Iyer et al. (Iyer et al., 2011). Labelled HAE and PCxxC motifs are highlighted in yellow and denoted with #.
