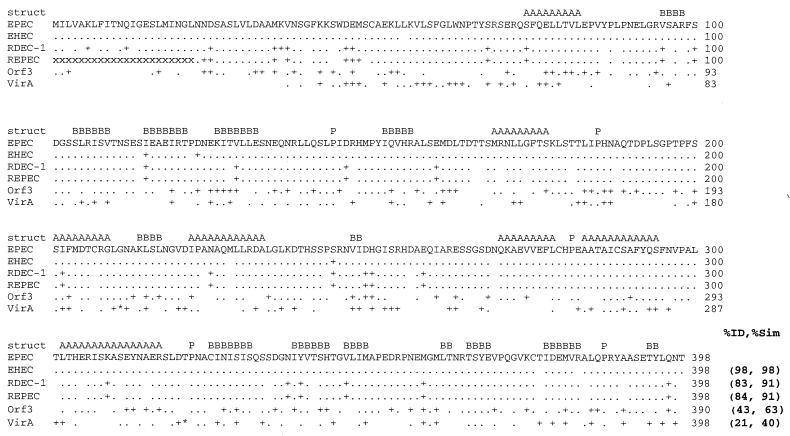
FIG. 1.
Alignment of EspG from EPEC O127: H7 (EPEC) with EspG from EHEC O157: H7 (EHEC), rabbit pathogen RDEC-1, REPEC strain 83/39 (REPEC), Orf3 from the EspC pathogenicity islet, and VirA from S. flexneri. Numbers at the end of the line, amino acid numbers; period, identical amino acids; +, similar but nonidentical amino acids; blank space, nonhomologous amino acids. The DNA sequence encoding the first 22 amino acids (aa) of REPEC VirA is not known, and each of these missing amino acids is indicated with an “x.” Asterisks (within the VirA sequence), areas where VirA contains extra amino acids not found within EspG, including an insertion of 5 aa into the region corresponding to aa 210 and 211 in EspG and a 15-aa insertion between aa 320 and 321 in EspG. The predicted consensus secondary structures (struct) conserved between EspG and VirA are denoted A (α-helix), B (β sheet), and P (proline; indicating a turn). The percent identity and percent similarity (%ID and %Sim, respectively) to the sequence of EPEC EspG are listed at the end of the alignment.