Fig 4: SD alters the reactivation of NRAM-GFP expressing neurons assessed by co-localization with cFos and Npas4.
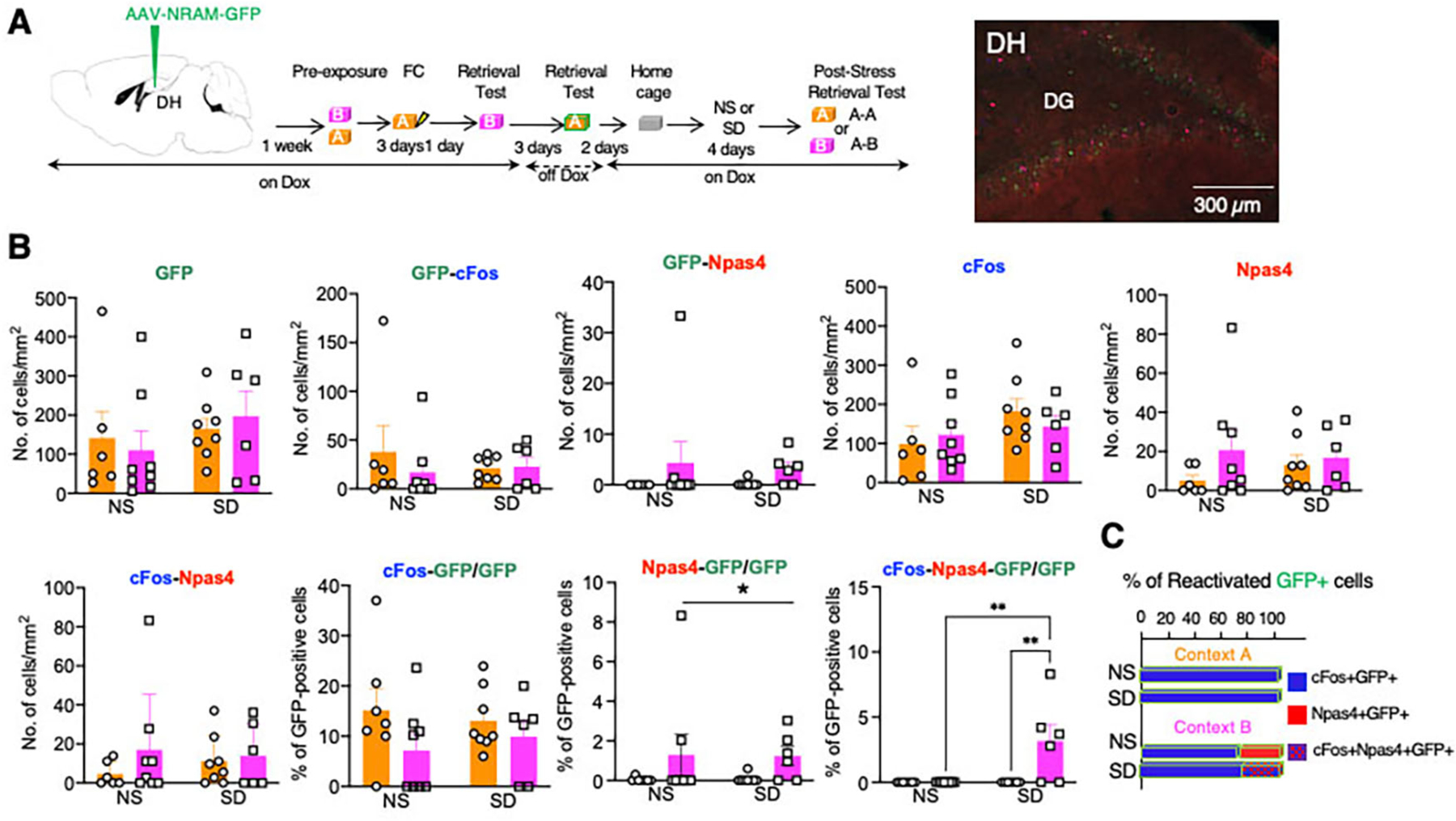
A Left, timeline of AAV9-N-RAM-d2tTA-TRE-GFP infusion, Dox regimen, and retrieval tests. As in the previous experiment, labeling of DH neurons with NRAM-GFP was performed at the first retrieval test in Context A, whereas reactivation was assessed after the second retrieval test either in Contexts A (A-A) or B (B-B). Middle, illustration of triple labeling of DG neurons with GFP, cFos, and Npas4. Right, illustration of GFP labeling in mice kept ON-Dox during the entire behavioral paradigm. B The number of GFP-, cFos-, and Npas4-positive DG neurons and their combinations after the post-stress retrieval test of SD mice or corresponding retrieval test of NS mice. The number of Npas4/GFP-co-labelled neurons was significantly increased after retrieval in Context B, whereas the number of triple labeled cFos/Npas4/GFP neurons was only detected in SD but not NS mice. *p < 0.05. Two-Way ANOVA. GFP: Stress F (1, 24) = 1.127, P=0.2990. Context F (1, 24) = 0.0001643, P=0.9899. Stress x Context F (1, 24) = 0.3764, P=0.5453. n = 6–8 mice in each group. GFP-cFos: Stress F (1, 24) = 0.1462, P=0.7056. Context F (1, 24) = 0.4782, P=0.4959. Stress x Context F (1, 24) = 0.6292, P=0.4354. n = 6–8 mice in each group. GFP-Npas4: Stress F (1, 24) = 0.0362, P=0.8506. Context F (1, 24) = 2.147, P=0.1558. Stress x Context F (1, 24) = 0.08050, P=0.7791. n = 6–8 mice in each group. cFos: Stress F (1, 24) = 2.324, P=0.1404. Context F (1, 24) = 0.05052, P=0.8241. Stress x Context F (1, 24) = 0.8190, P=0.3745. n = 6–8 mice in each group. Npas4: Stress F (1, 24) = 0.08408, P=0.7743. Context F (1, 24) = 1.762, P=0.1968. Stress x Context F (1, 24) = 0.6845, P=0.4162. n = 6–8 mice in each group. cFos-Npas4: Stress F (1, 24) = 0.05915, P=0.8099. Context F (1, 24) = 1.111, P=0.3024. Stress x Context F (1, 24) = 0.4367, P=0.5024. n = 6–8 mice in each group. cFos-GFP/GFP: Stress F (1, 25) = 0.01053, P=0.9191. Context F (1, 25) = 2.917, P=0.1000. Stress x Context F (1, 25) = 0.5415, P=0.4687. n = 6–8 mice in each group. Npas4-GFP/GFP: Stress F (1, 25) = 0.02197, P=0.8834. Context F (1, 25) = 4.470, P=0.0446. Stress x Context F (1, 25) = 0.04054, P=0.8421. n = 6–8 mice in each group. cFos-Npas4-GFP/GFP: Stress F (1, 24) = 8.682, P=0.0070. Context F (1, 24) = 8.682, P=0.0070. Stress x Context F (1, 24) = 8.682, P=0.0070, n = 6–8 mice in each group. C In the A-A condition, all reactivated GFP neurons were positive for cFos, whereas in the A-B condition 18–20% of the reactivated neurons were Npas4-positive. Depending on the stress condition, the Npas4/GFP neurons were either a separate neuronal population (NS group) or a population co-labeled with cFos (SD).
