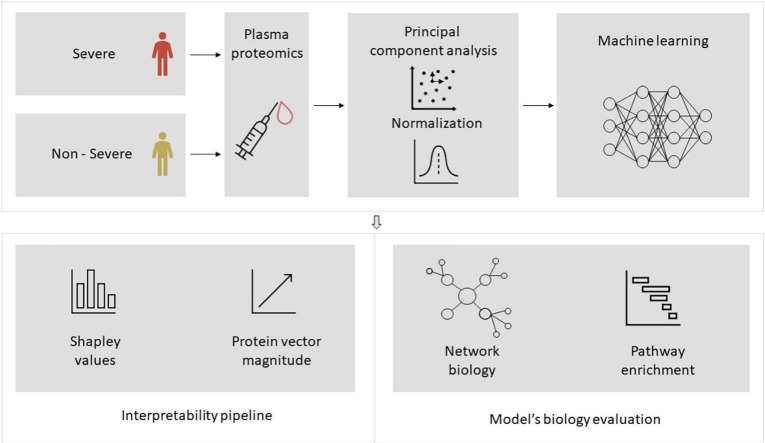
Abstract
The SARS-CoV-2 pandemic highlighted the need for software tools that could facilitate patient triage regarding potential disease severity or even death. In this article, an ensemble of Machine Learning (ML) algorithms is evaluated in terms of predicting the severity of their condition using plasma proteomics and clinical data as input.
An overview of AI-based technical developments to support COVID-19 patient management is presented outlining the landscape of relevant technical developments. Based on this review, the use of an ensemble of ML algorithms that analyze clinical and biological data (i.e., plasma proteomics) of COVID-19 patients is designed and deployed to evaluate the potential use of AI for early COVID-19 patient triage. The proposed pipeline is evaluated using three publicly available datasets for training and testing. Three ML “tasks” are defined, and several algorithms are tested through a hyperparameter tuning method to identify the highest-performance models. As overfitting is one of the typical pitfalls for such approaches (mainly due to the size of the training/validation datasets), a variety of evaluation metrics are used to mitigate this risk.
In the evaluation procedure, recall scores ranged from 0.6 to 0.74 and F1-score from 0.62 to 0.75. The best performance is observed via Multi-Layer Perceptron (MLP) and Support Vector Machines (SVM) algorithms. Additionally, input data (proteomics and clinical data) were ranked based on corresponding Shapley additive explanation (SHAP) values and evaluated for their prognosticated capacity and immuno-biological credence. This “interpretable” approach revealed that our ML models could discern critical COVID-19 cases predominantly based on patient's age and plasma proteins on B cell dysfunction, hyper-activation of inflammatory pathways like Toll-like receptors, and hypo-activation of developmental and immune pathways like SCF/c-Kit signaling. Finally, the herein computational workflow is corroborated in an independent dataset and MLP superiority along with the implication of the abovementioned predictive biological pathways are corroborated.
Regarding limitations of the presented ML pipeline, the datasets used in this study contain less than 1000 observations and a significant number of input features hence constituting a high-dimensional low-sample (HDLS) dataset which could be sensitive to overfitting. An advantage of the proposed pipeline is that it combines biological data (plasma proteomics) with clinical-phenotypic data. Thus, in principle, the presented approach could enable patient triage in a timely fashion if used on already trained models. However, larger datasets and further systematic validation are needed to confirm the potential clinical value of this approach. The code is available on Github: https://github.com/inab-certh/Predicting-COVID-19-severity-through-interpretable-AI-analysis-of-plasma-proteomics.
Keywords: COVID-19, Artificial intelligence, Machine Learning, Forecasting, Severity prediction
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has caused a global pandemic (COVID-19) leading to millions of deaths worldwide [1]. COVID-19 has a highly heterogenous clinical course, ranging from asymptomatic patients or mild symptoms to severe pneumonia with acute respiratory syndrome (ARDS) that frequently leads to death. Although vaccination efforts are ongoing worldwide with significant impact, the emergence of novel mutated Sars-CoV-2 variants constantly poses new healthcare challenges [2]. Since the COVID-19 pandemic constitutes a significant economic burden on public health systems worldwide, ongoing efforts for novel biomarker discovery based on high-throughput -omic technologies to improve patient triage are of obvious scientific and public health interest.
In the era of “Big Data” in healthcare, Artificial Intelligence (AI) and Machine Learning (ML) are heavily investigated for the development of diagnostic, prognostic and predictive supervised models [3], [4], [5]. While ML is a promising technical paradigm, still, it also poses several challenges and often its results produced “in silico” are difficult to validate in real-world healthcare settings. To this end, standardization, benchmarking and validation of AI/ML approaches have been identified as critical open issues to promote “trust” on the produced models.
Initially, this study presents an overview of the application of ML for COVID-19 patient stratification based on their condition severity focusing on proteomic data. Next, based on the findings in the literature, a computational pipeline is proposed, and a benchmarking analysis of the employed ML algorithms is presented organized in three distinct ML tasks. The pipeline reveals that Multi-Layer Perceptron (MLP) outperforms other common ML classifiers, by prioritizing plasma proteins that participate in known and not insofar explored biological pathways, hence highlighting its translational significance. Overall, the herein approach tries to address the “black box” challenge of ML approaches [6] in the analysis of -omic data from multi-factorial diseases like COVID-19 with a combination of “interpretable” AI methodologies [7] and outlines a computational methodology1 for the investigation of potential biomarkers that could help carefully stratify patients with unfavorable clinical trajectories.
2. Assessing current literature
Several papers investigating the use of ML to support the management of COVID-19 patients have been published. However, only a few papers use AI upon proteomic data for COVID-19. In this section, we outline relevant AI-based analysis pipelines on Olink NPX technology proteomic data, organized in the typical stages of ML pipelines, i.e., data preprocessing, training and interpretation.
2.1. Data preprocessing
In the prediction models, a preprocessing pipeline is followed to face the problem of data gaps or errors. The two most prominent approaches were the deletion of records that did not satisfy the quality criteria and/or the imputation of the null values, i.e., their replacement with “proper” values that would facilitate computations but not introduce significant biases. From our research, the following imputation approaches are identified:
-
-
Multiple imputation method using Fully Conditional Specification (FCS) [10]: The missing values are imputed based on the observed values for a given individual and the relations observed in the data for other participants, assuming the observed variables are included in the imputation model.
-
-
k - nearest neighbors (k-NN) imputation [8]: This method selects observations with similar characteristics to the observation of interest to impute missing values (e.g., based on the use of Euclidean distance between observations as a similarity measure).
Sometimes, input data are normalized (e.g. at the study of Gisby et al. [8], data is standardized with a mean of 0 and standard deviation of 1). Moreover, several methods were employed to feature selection and/or dimensionality reduction. Table 1 identifies the different approaches applied in the COVID-19 proteomic data preprocessing.
Table 1.
Feature selection/dimensionality reduction approaches.
| Dimensionality reduction methods | Description |
|---|---|
| Recursive feature extraction (RFE) analysis [9] | Eliminates the features that less correlated with the target variable. |
| Feature selection through GINI Index [9] | The features are selected based on GINI Index, a measure that calculates the contribution of each feature to the prediction of the output. |
| Recursive Feature Elimination [10] | It is a feature importance method that keeps the most significant features for the chosen ML model. |
| Principal Component Analysis (PCA) [10] | It is a dimensionality reduction process that integrates variables of a dataset to uncorrelated features and maximizes variance. |
2.2. ML training approaches
Most of the reviewed works, applied ensembles of ML algorithms, typically including Random Forest, Gradient Boosted Decision Tree, XGBoost, and Extra Tree classifiers. Logistic regression, Lasso Logistic regression, Support Vector Machine (SVM), and neural network algorithms were also individually applied in some papers. A notable contribution was proposed by Byeon al. [11], who introduced the concept of an AutoML classifier (AutoGluon-Tabular).
2.3. Interpretability
While this was not the typical case, some works also focus on the respective models' interpretability. Indicatively, Shapley additive explanation (SHAP) values were used by Beltrami et al. [9], to depict the impact of the training features in the outcome model. Moreover, coefficients are calculated as an explanation method as the mean decrease in the Gini method [12] and minimal-optimal variables method [13]. Gisby et al. [8] applied a random forest explainer from an R library.
3. Methods
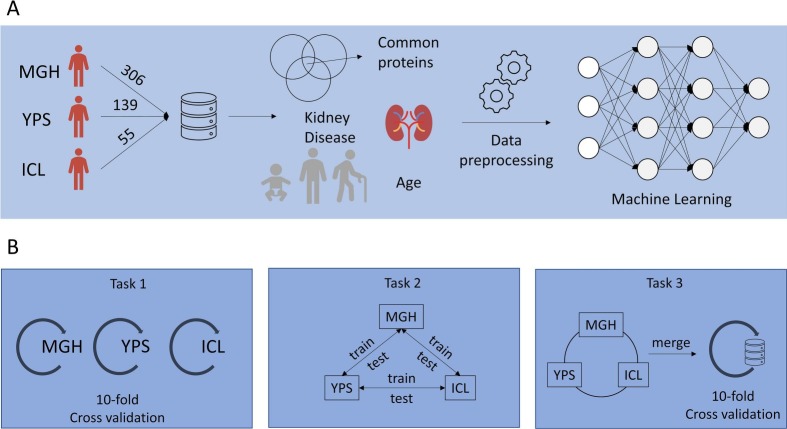
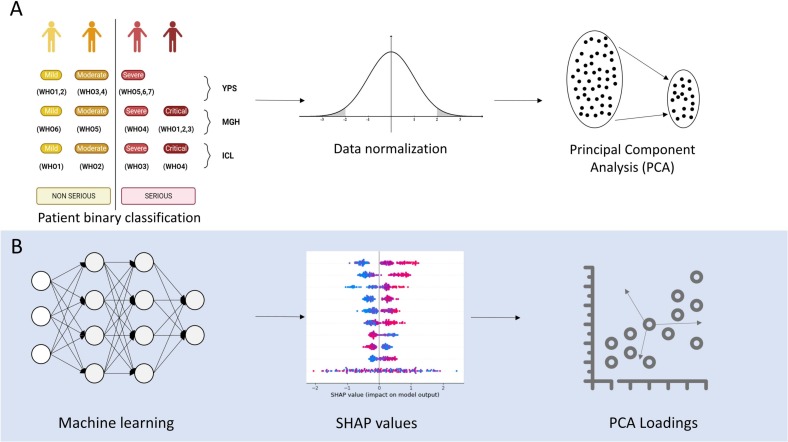
Three publicly available datasets were aligned to be used for training ( Fig. 1 – upper part A) and three ML “tasks” were defined to validate the application of the selected algorithms to evaluate their performance ( Fig. 1 – lower part B).
Fig. 1.
Methodology (part A: The overall approach of the methodology, part B: The Machine Learning tasks).
3.1. ML prediction tasks
The ML tasks can be summarized as follows:
-
-
In task 1, an ensemble of ML models was trained independently on each of the three datasets and validated with the cross-validation method using data from the same dataset. For this task, every dataset contained all the initial columns/features.
-
-
In task 2, each dataset was used for training, and the trained model was tested against the two remaining datasets. More specifically, for the training process, a 10-fold cross-validation procedure is used and then the trained models are tested separately against the rest of the datasets.
-
-
In task 3, the three datasets are merged in a great bulk dataset, and models are trained and tested with a 10-fold cross-validation method against data from the aggregated bulk dataset.
The ML models for the three tasks were developed following the TRIPOD (Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis) guidelines (Supplementary Fig. 12) [14].
Supplementary Fig. 12.
Evaluation task performance from GridSearch hyperparameter tuning.
3.2. Data collection
The first dataset originated from the seminal study of the Massachusetts General Hospital (MGH) and contained 306 COVID-19 patients [15], the second dataset originates in the Imperial College of London (ICL) and involved 55 End-Stage Renal Disease (ESRD) patients [16], and the third dataset comes from the Institute for Systems Biology of Seattle (YPS) consisting of 139 COVID-19 patients [17]. The selected datasets contain plasma proteomic data and include information about patients' comorbidities, clinical profiles, symptoms, demographics, other -omic data, and prognosis scores based on World Health Organization (WHO) grading scheme [18]. Population characteristics for the study cohorts are presented in Table 2 . Interestingly, a common clinical denominator among the three studies in focus was kidney disease comorbidity, since the latter was strongly associated with poor clinical prognosis [19].
Table 2.
Population characteristics for the study cohorts.
| COVID-19 severity outcome | MGH |
YPS |
ICL |
|||
|---|---|---|---|---|---|---|
| Severe (n = 132) |
Non-severe (n = 251) |
Severe (n = 50) |
Non-severe (n = 206) |
Severe (n = 13) |
Non-severe (n = 42) |
|
| Age, years range | ||||||
| (20,40] | 5 (3.8 %) | 31 (12.3 %) | 6 (12 %) | 62 (30 %) | 0 | 2 (4.8 %) |
| (40,60] | 16 (12.1 %) | 57 (23 %) | 15 (30 %) | 52 (25.2 %) | 1 (7.7 %) | 6 (14.3 %) |
| (60,80] | 80 (60.6 %) | 129 (51.4 %) | 25 (50 %) | 64 (31 %) | 9 (69.2 %) | 26 (62 %) |
| (80,100] | 31 (23.5 %) | 34 (13.5 %) | 4 (8 %) | 28 (13.6 %) | 3 (23 %) | 8 (19 %) |
| Kidney disease | ||||||
| Positive | 30 (22.7 %) | 31 (12.3 %) | 4 (8 %) | 14 (6.8 %) | 13 | 42 |
| Negative | 102 (77.3 %) | 220 (87.7 %) | 46 (92 %) | 192 (93.2 %) | 0 | 0 |
3.3. Data pre-processing and feature selection
The three datasets were normalized and aligned so that they could be used both individually (task 1 and task 2) and also combined in one (task 3). Proteomics data, the age category of patients, and characterization of kidney disease comorbidity were used as the input features of our approach, presented in detail in Table 2.
It should be noted that the alignment process of the three datasets for task 2 and task 3 led to the removal of data columns that refer to data not common between the three datasets. Before alignment, three rows that contained almost 10% of null values were detected and removed in YPS dataset. Additionally, the null column of the CD6 protein was deleted. Similarly, null values of MGH and ICL datasets were deleted for the first task. For task 1, 33 patients with null values were deleted from the MGH dataset, and two patients from the ICL dataset. In tasks 2 and 3, there are no null values in the common columns that are used for the ML models.
To align the data provided by the three datasets, we categorized the age values into four main categories based on the initial separation of the MGH dataset ((20,40] = 1, (40,60] = 2, (60,80] = 3, (80,100] = 4). Furthermore, as all ICL patients were characterized as positive for kidney disease comorbidity in the paper, there is no representative column. Thus, a binary column was added (value 1).
Moreover, as the expression of proteins in every dataset is provided in arbitrary Normalized Protein eXpression (NPX) units [1] with a distinct range of values, the respective values are normalized using normal distribution applied to every dataset separately, using the StandardScaler tool [20]. Furthermore, the COVID-19 severity outcome column is transformed to binary, corresponding to “Serious” and “Non-serious” based on the WHO score as described in Fig. 3A. Table 3 presents a complete overview of the feature structure in our datasets.
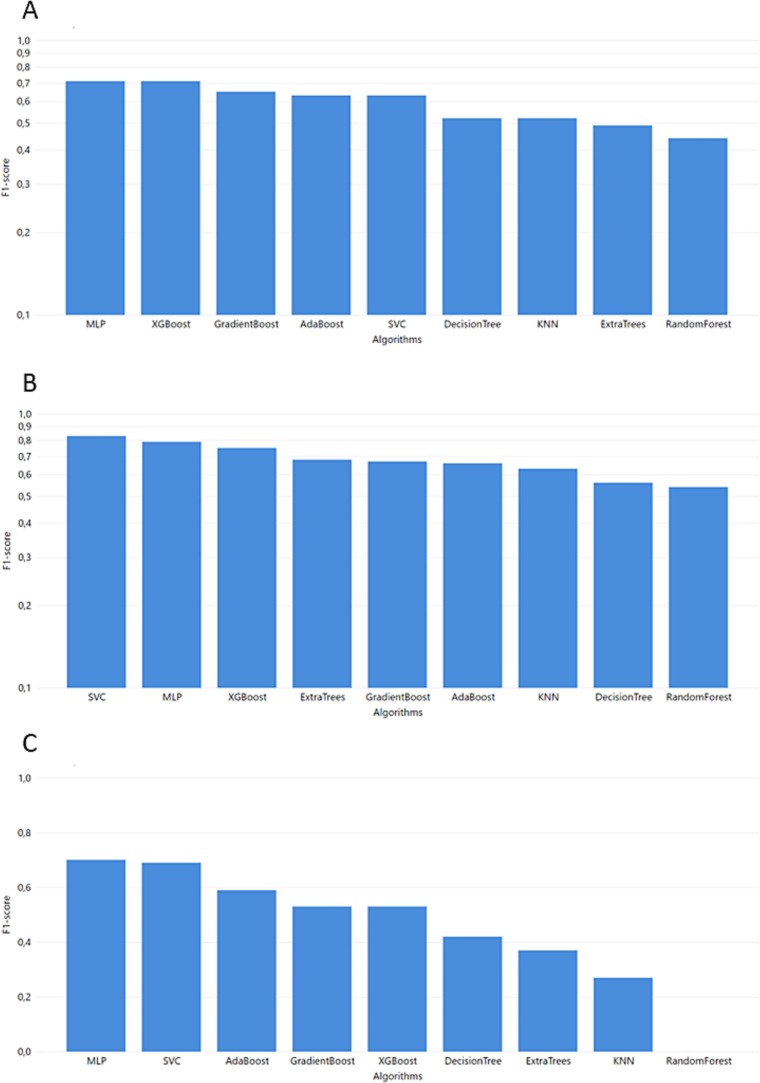
Fig. 3.
Task 1 performance of F1-score from GridSearch hyperparameter tuning A. MGH B. YPS C. ICL.
Table 3.
Description of the common features of the 3 datasets.
| Features | Description |
|---|---|
| Proteins | An intersection of 168 common proteins identified in the three selected datasets (Supplementary Fig. 1). Protein expression is measured in NPX (Normalized Protein eXpression) which is Olink's arbitrary unit and it is normalized in Log2 scale. Sequentially, the values were standardized in a range from 0 to 1. |
| Age | Patients are separated in the following classes: (20,40], (40,60], (60,80], (80,100] |
| Kidney disease comorbidity | Dataset demarcated patients over kidney disease. |
| COVID-19 patients' severity | Patients are categorized based on the WHO Progression scale as Severe COVID-19 (severe, critical) and Non-severe COVID-19 (mild, moderate) [21]. |
Regarding the dimensionality reduction approach applied, Tsai et al. [22] argue that the selection of highly sophisticated dimensionality reduction algorithms might not have a significant effect on High Dimensionality and Low Sample (HDLS) datasets (such as the ones used in the presented pipeline). Thus, the dimensionality reduction method applied was based on the Principal Component Analysis (PCA) approach (Fig. 2A). More specifically, PCA was applied only to the protein expression columns which corresponded to different dimensions for the different trained datasets (Table 4 ):
Fig. 2.
Development and evaluation of the final models A Data preprocessing B Feature contribution on the models' results.
Table 4.
Dimensionality reduction with PCA.
| Tasks | Datasets | Initial dimensions | Final dimensions |
|---|---|---|---|
| Task 1 | MGH | 1420 | 124 |
| YPS | 457 | 66 | |
| ICL | 438 | 35 | |
| Task 2 | MGH | 168 | 61 |
| YPS | 168 | 48 | |
| ICL | 168 | 30 | |
| Task 3 | MGH- YPS- ICL | 168 | 72 |
3.4. Machine Learning algorithms
For our experiment, several ML algorithms are tested for their performance at the prediction tasks mentioned above.
3.4.1. K-nearest neighbors (KNN)
KNN is a classification learning algorithm that is placing the training sample in an n-dimensional space. Next, every observation from test dataset is classified (mostly based on the Euclidean distance) with the training samples that is closer to [23].
3.4.2. Decision tree
A decision tree algorithm is based on a tree-like structure, where each test feature is represented by an internal node, where every classification is represented by each leaf [24].
3.4.3. Random Forest
Random Forest is a combination of decision trees, that can be used for both classification and regression. It is an ensemble-based ML method [25].
3.4.4. Support Vector Machine (SVM)
The SVM modeling algorithm is seeking for the optimal hyperplane that distinguish two classes with the greatest margin [26]. This demands to solve the following maximization problem (1):
| (1) |
3.4.5. Extra Trees
Extra Trees algorithm builds totally randomized trees whose structures are independent of the output values of the learning sample. It is an ensemble-based ML method [27].
3.4.6. Extreme gradient boosting (XGBoost)
XGBoost is a regression tree, and it can apply regression and classification. This algorithm is a variant of the gradient boosting machine (GBM) [28]. If the XGBoost model consists of K decision trees, the optimization objective function is the below Eq. (2):
| (2) |
fk: independent tree with leaf scores
F: space of regression tree
3.4.7. Multi-Layer Perceptron (MLP)
The MLP is a fully connected feedforward artificial neural network. An MLP consists of input nodes that connect as a directed graph with the nodes of the output layer [29]. It is a supervised learning algorithm that learns a function (3):
| (3) |
m: number of input's dimensions
o: number of output's dimensions
3.5. Evaluation metrics
The accuracy of KNN, Decision Tree, Random Forest, SVM, Extra Trees, XGBoost, and MLP classifiers were tested for all the above three tasks. The selection of the model parameters was performed via the grid search method for each ML algorithm through cross-validation. Thus, the combination of parameters chosen for the models at every task is presented as supplementary material along with the respective results for every evaluation metric (accuracy, area under the curve - AUC, precision, recall, and F1-score).
3.6. Interpretation of ML results through Systems Biology
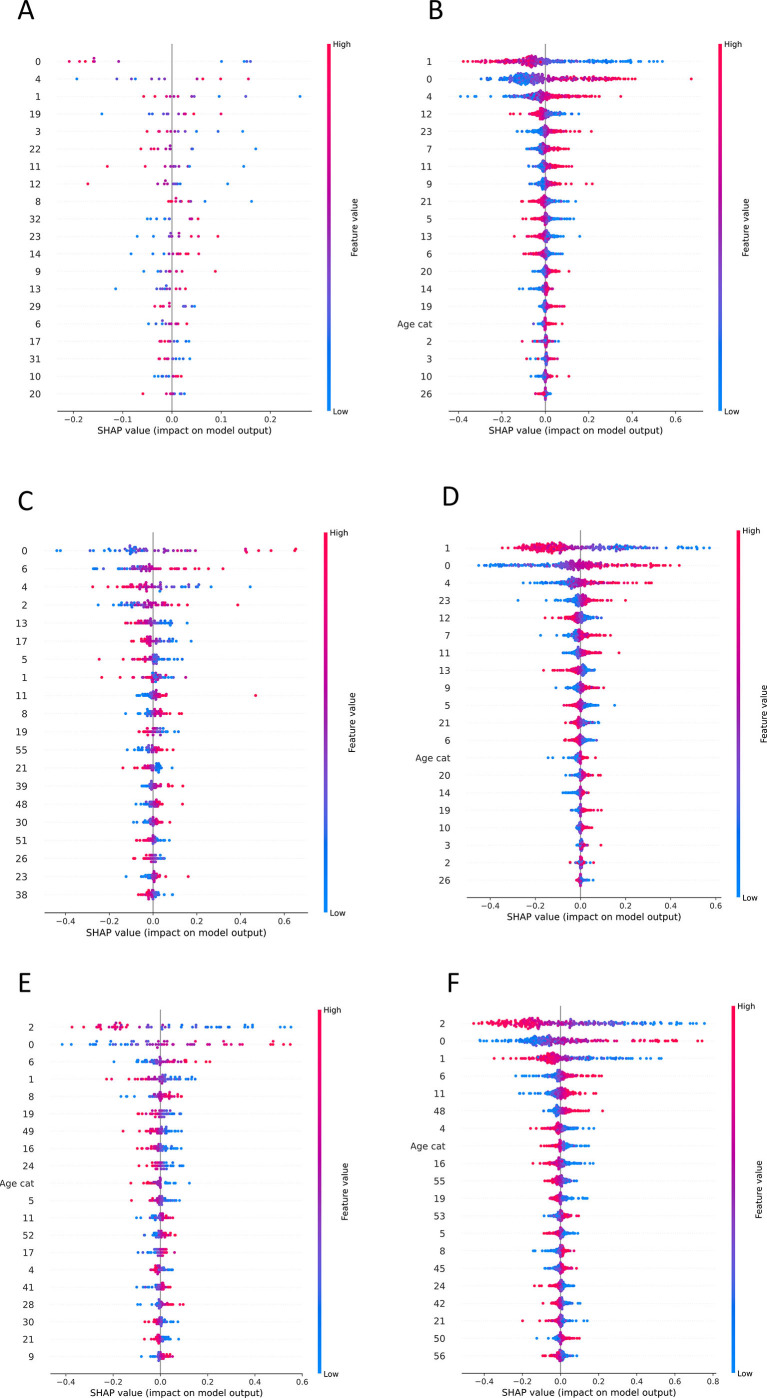
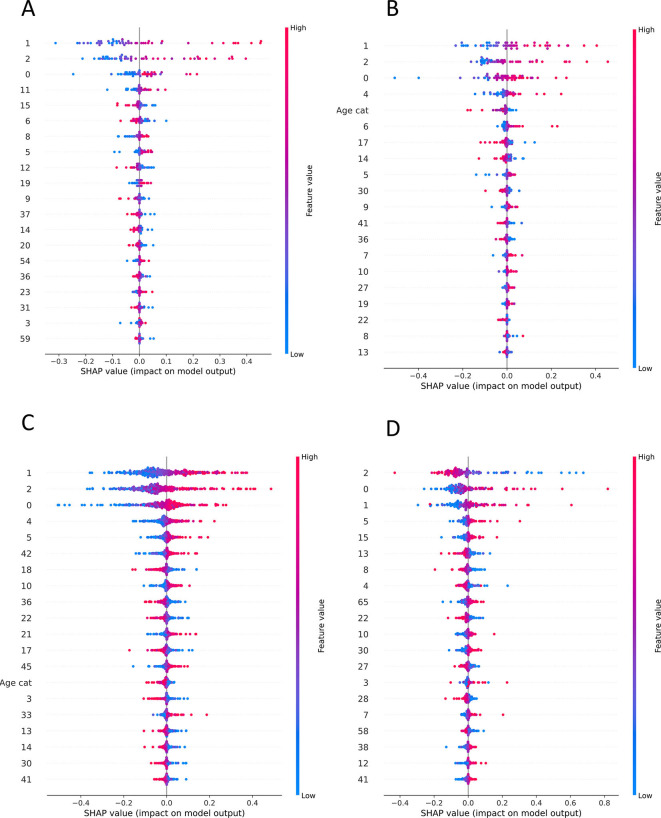
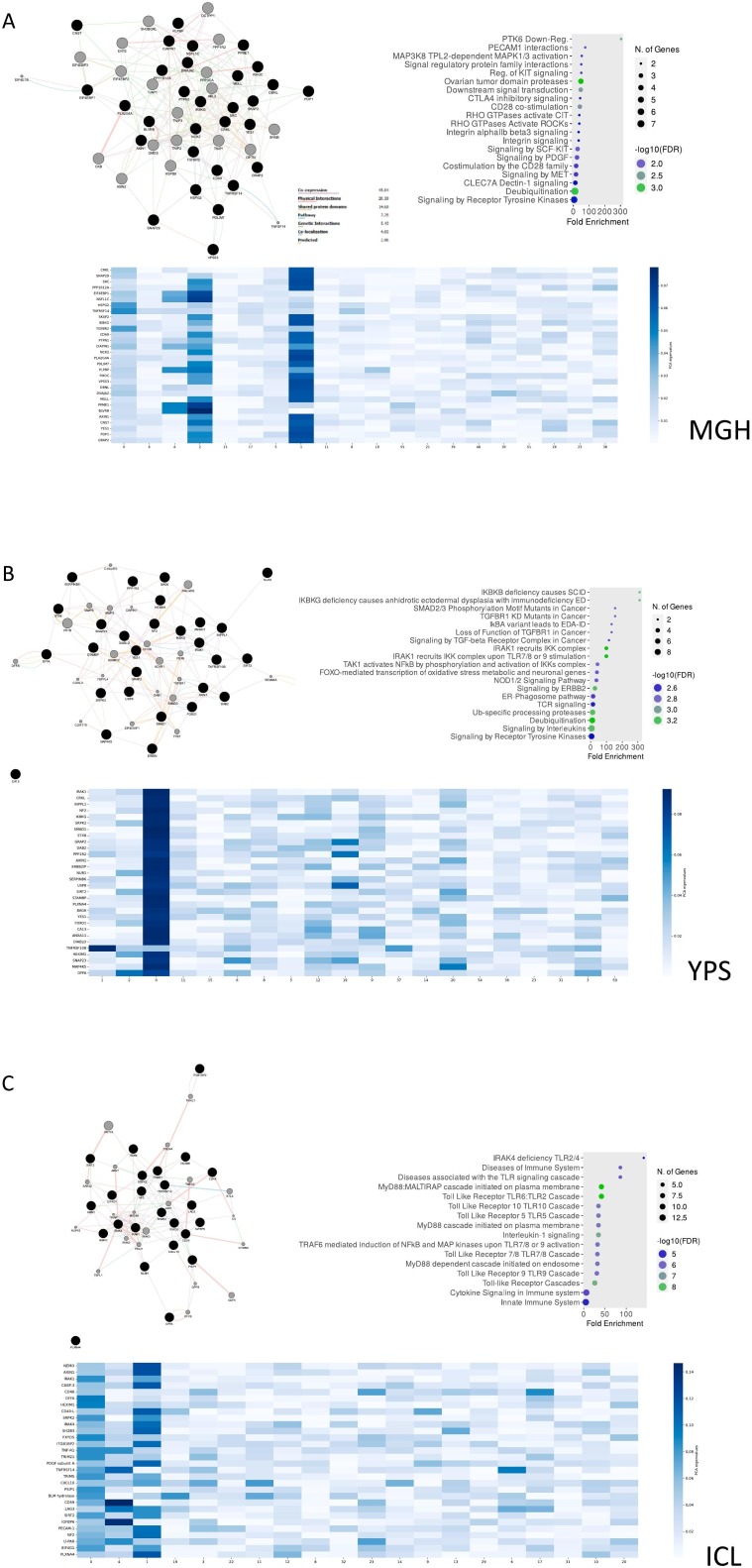
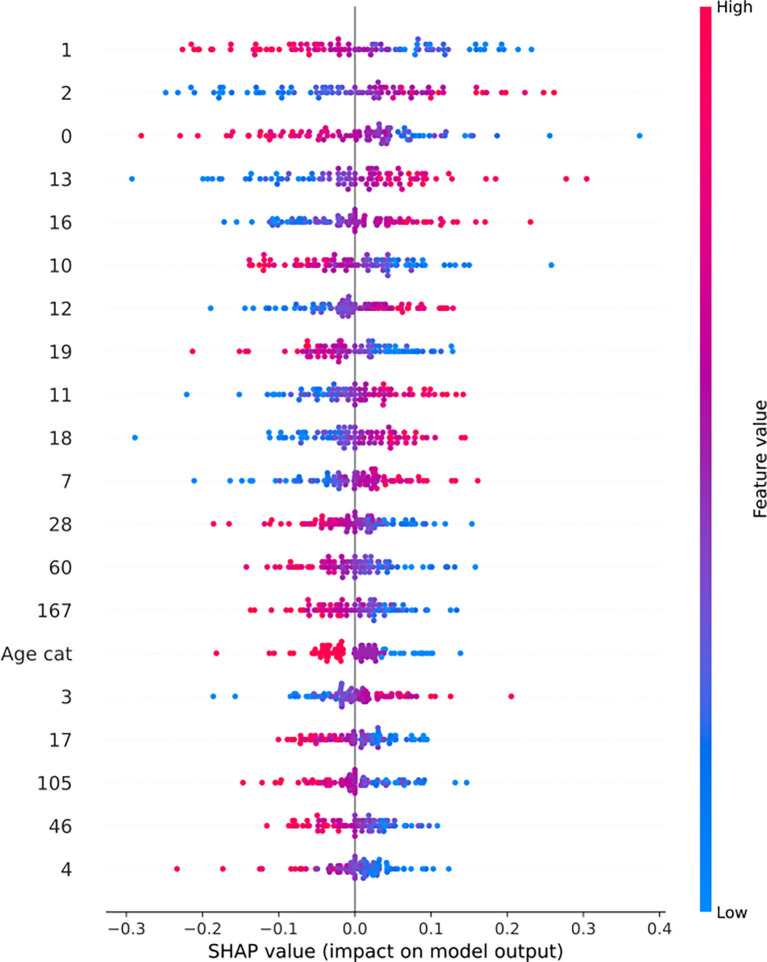
To evaluate the produced results, SHAP values were calculated for every feature as they came up after the PCA application (Supplementary Figs. 6, 7). As protein expression features are represented via vectors after the dimensionality reduction of PCA, we computed the loadings for every component that was in the top 20 contributors of the predictive model result. Furthermore, the magnitude of every protein from these components is calculated (Fig. 3B). Finally, to gain a better understanding of the biological interpretability of the produced ML models, we also performed pathway enrichment approaches using Enrichr (“Reactome” database) for the plasma proteins ranked as the top-30 loadings (Supplementary Figs. 8, 9, 10) and constructed protein-protein interaction networks (PPI) with GeneMania to unravel biological interactions among plasma proteins of interest [30].
Supplementary Fig. 6.
SHAP values of ML models that are presented the contribution of PCA components to the final classification. A. SHAP values of Task 1 MLP model trained with ICL dataset B. SHAP values of Task 2 SVM model trained with ICL dataset and tested to MGH C. SHAP values of Task 2 SVM model trained with ICL dataset and tested to YPS D. SHAP values of Task 1 MLP model trained with MGH dataset E. SHAP values of Task 2 MLP model trained with MGH dataset and tested to ICL F. SHAP values of Task 2 MLP model trained with MGH dataset and tested to YPS.
Supplementary Fig. 7.
SHAP values of ML models that are presented the contribution of PCA components to the final classification. A. SHAP values of Task 1 MLP model trained with YPS dataset B. SHAP values of Task 2 MLP model trained with YPS dataset and tested to ICL C. SHAP values of Task 2 MLP model trained with YPS dataset and tested to MGH D. SHAP values of Task 3 SVM model.
Supplementary Fig. 10.
MLP predicts COVID-19 severity in the independent MC dataset, using similar proteins and pathways to MGH-YPS-ICL datasets. GeneMania protein-protein interaction networks (PPI), pathway enrichment dotplots and heatmaps with protein-nodes for the most predictive plasma proteins (top-30) as features of MLP (Task 1) considering for the validation dataset MC.
4. Results
The grid-search cross-validation approach led to the results of the best parameters with the highest accuracy of each one of the three tasks. The performance of the respective ML models was presented using AUC, F1-score, Precision, and Recall. It should be noted that as the classification of severe COVID-19 patients is more important than the early identification of patients with mild prognosis, the ML models are selected based on the recall of severe COVID-19 patients.
4.1. Task 1
The performance from the first task was separated into three parts for the three different datasets.
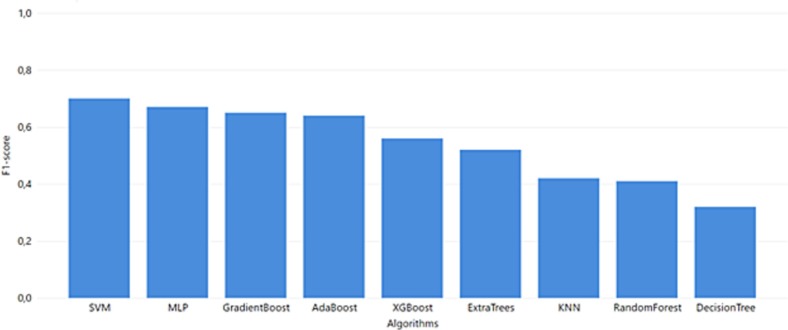
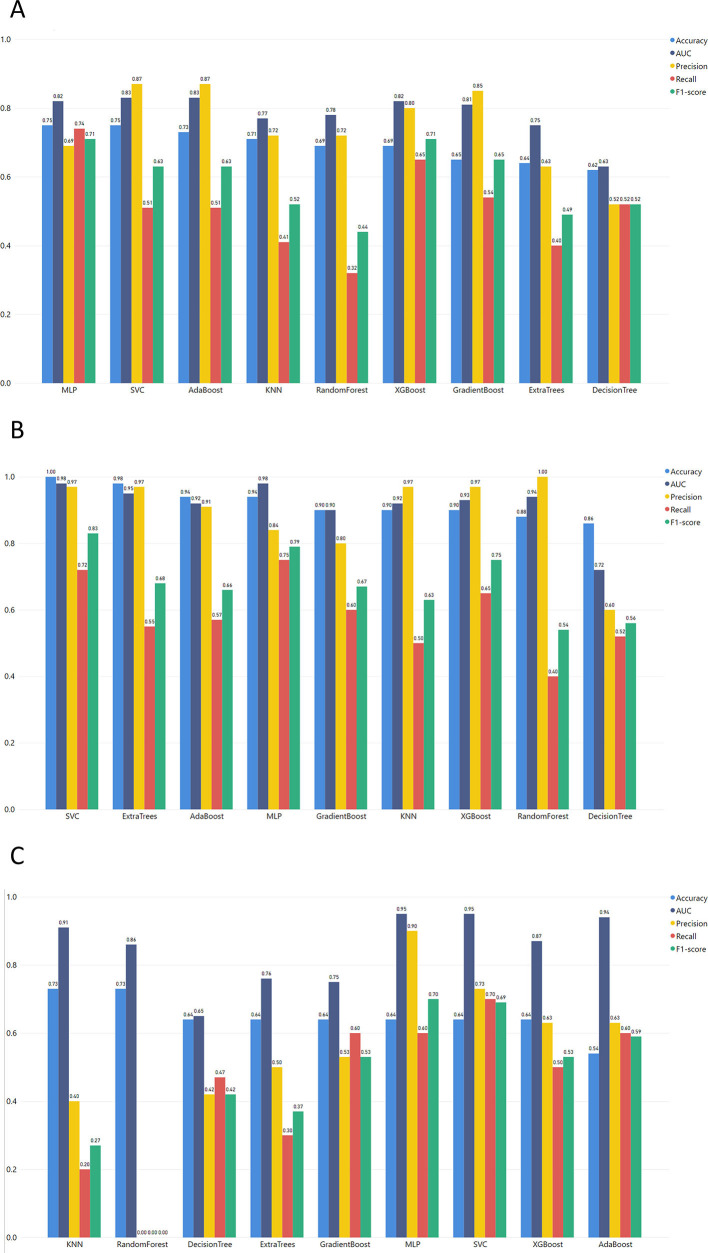
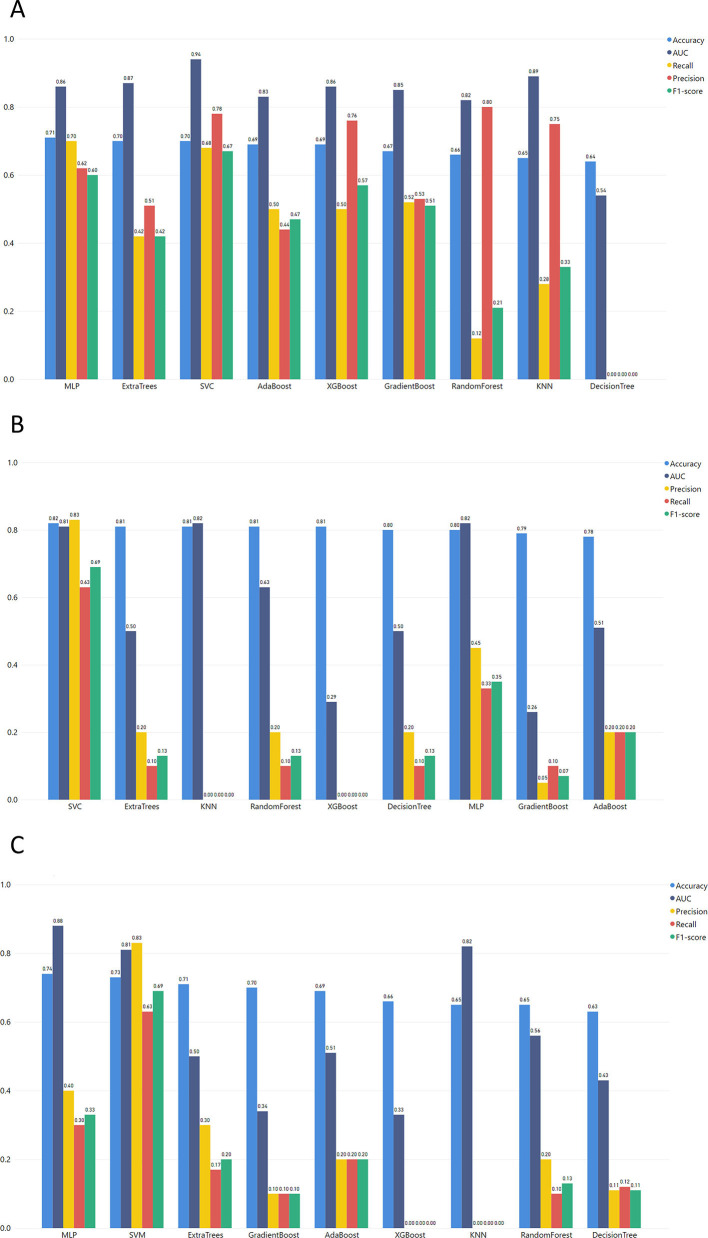
The best ML model for the MGH dataset (Fig. 3) was achieved via MLP which combines the best performance in accuracy, F1-score, and recall with 0.75, 0.71, and 0.74, respectively. Additionally, MLP succeeded in one of the highest AUC scores. The precision of the MLP model was lower than most of the models but for the specific use case, false positives (non-severe cases falsely predicted as severe) are preferable to false negatives (unpredicted severe COVID-19 cases). Thus, a high recall score is more important as a metric, and we prioritize it when evaluating the produced outcomes.
Regarding the YPS dataset (Fig. 4 ), high accuracy, AUC, and Precision scores were achieved with the highest F1-score, and recall succeeded from computationally complex algorithms like SVM reached 0.83 and 0.72, respectively. MLP and SVM algorithms are the highest performers in every evaluation metric. However, these algorithms are sensitive to potential overfitting trained with small datasets (SVM accuracy 1.0 and MLP accuracy 0.94). Therefore, the XGBoost algorithm (an ensemble method for avoiding overfitting in small datasets) could be identified as a non-overfitting alternative (F1 score 0.75, recall 0.65).
Fig. 4.
Task 2 performance of F1-score from GridSearch hyperparameter tuning for trained model in MGH dataset and test in ICL and YPS dataset A. YPS B. ICL.
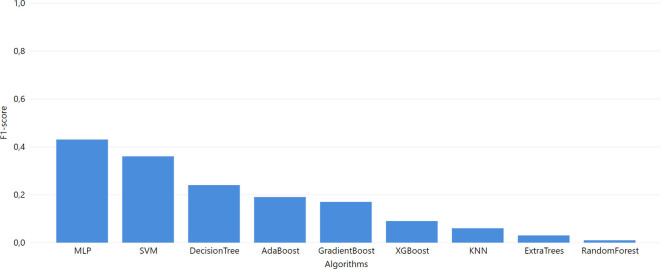
As we can see from the overall metrics (Fig. 4), the developed ML models that were applied to the ICL dataset did not perform very well. Since it is more important for our purpose to have the highest values for Recall and F1-score, the SVM algorithm seems to be the best choice (Recall = 0.7 and F1-score = 0.69).
4.2. Task 2
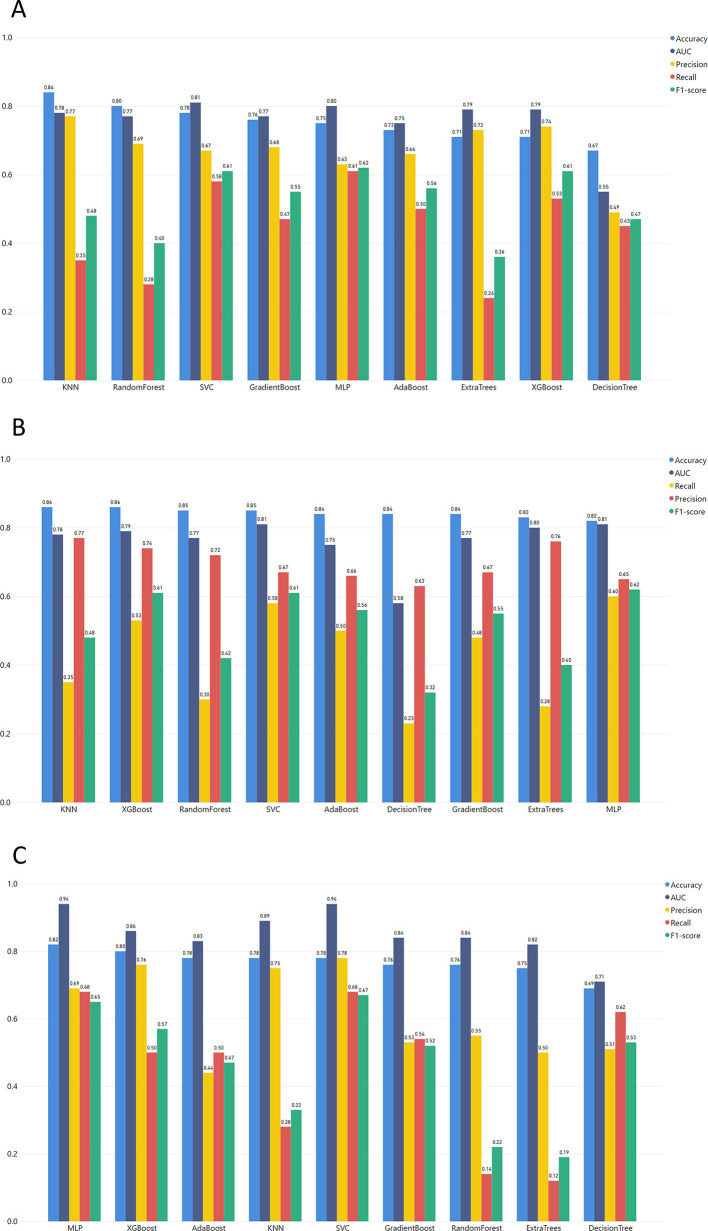
Regarding this task, it is significant to mention that the training models are more robust because the training and the testing sets come from different studies' datasets.
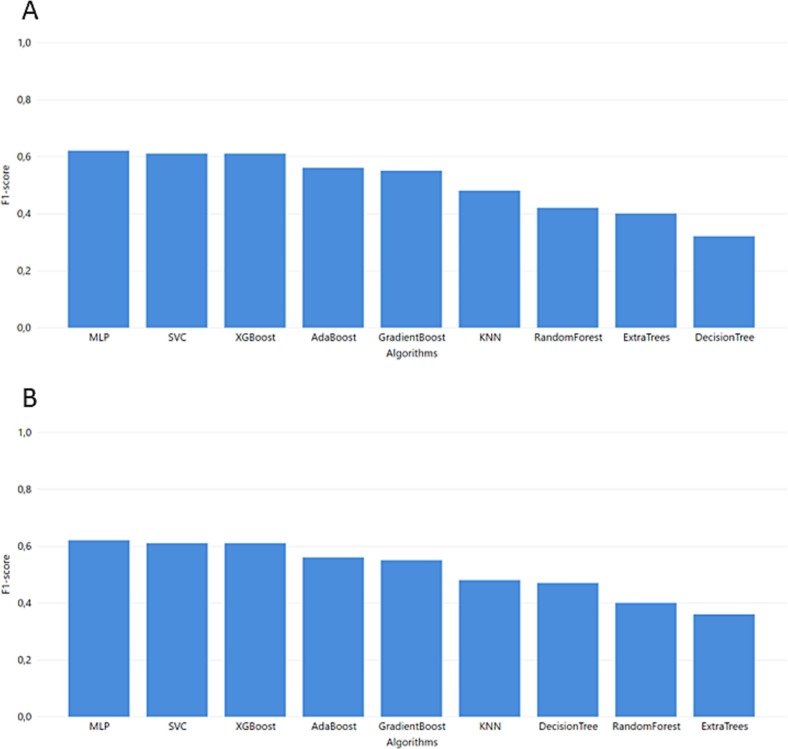
In the first step, the ML model was trained in the MGH dataset and tested in the ICL dataset (Fig. 4). In this context, the MLP algorithm achieved the highest F1-score and recall, with 0.62 and 0.61, respectively. Additionally, Fig. 5 depicts the performance of the models trained in the MGH dataset and tested using the YPS dataset, with MLP achieving the best performance.
Fig. 5.
Task 2 performance of F1-score from GridSearch hyperparameter tuning for trained model in YPS dataset and test in ICL and MGH dataset A. ICL B. MGH.
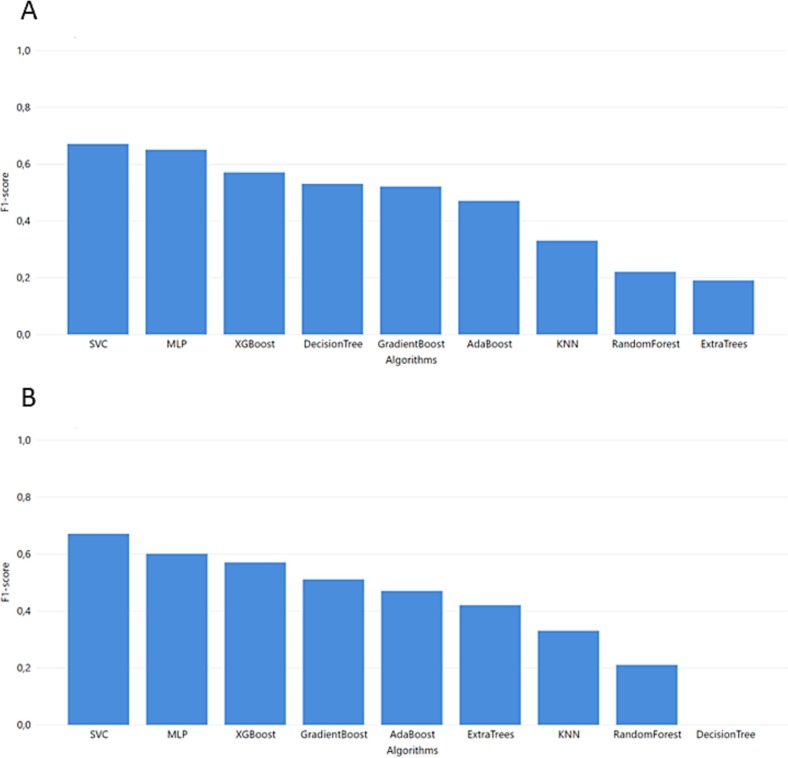
The models trained with the YPS dataset and tested with the MGH dataset (Fig. 5) produced relatively good results for both SVM and MLP models with slight differences in the recall (SVM reached 0.68 and MLP 0.7). On the other hand, there is a significant difference in precision between SVM (0.78) and MLP (0.62). To this end, the F1-score of MLP (0.6) is lower than SVM (0.67). As recall is the most important metric for our study, we conclude that MLP has the best performance in the correct classification of severe COVID-19 patients.
In Task 2, the last models were trained with the ICL dataset. Initially, these models were tested with the YPS dataset. As clearly shown in Fig. 6 , the SVM exceeded the rest of the models. The SVM model also outperforms the rest of the models when using the ICL dataset for training and MGH for validation (F1-score of 0.69).
Fig. 6.
Task 2 performance of F1-score from GridSearch hyperparameter tuning for trained model in ICL dataset and test in YPS and MGH dataset A. YPS B. MGH.
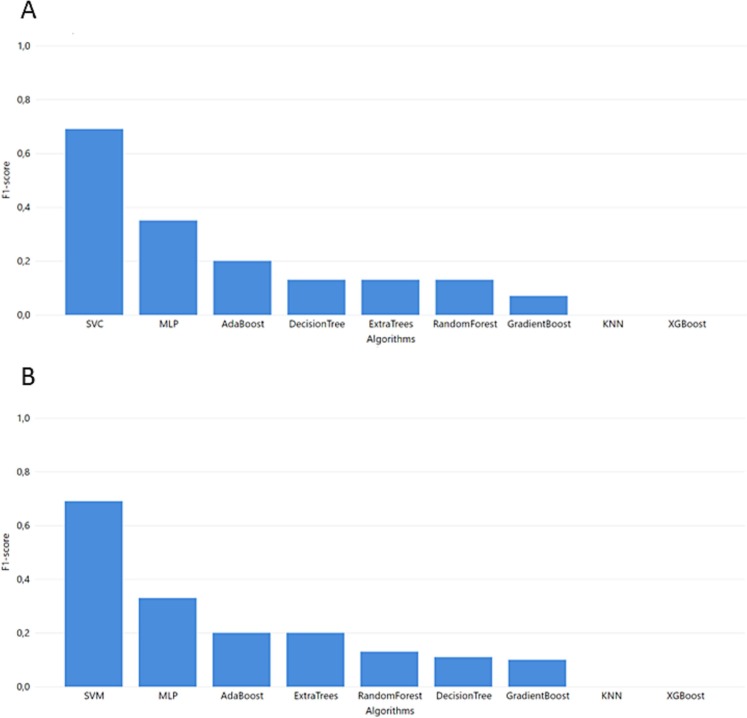
4.3. Task 3
For task 3 where all data are harmonized in one bulk dataset, SVM produced the best results (Fig. 7 ). Although in precision SVM reaches 0.77, the ensemble methods Gradient Boost, Random Forest, and Extra Trees surpass it with a precision of 0.78, 0.88, and 0.85, respectively (Supplementary Table 10).
Fig. 7.
Task 3 performance of F1-score from GridSearch hyperparameter tuning.
As a whole, the results from the trained models in Tasks 1 and 2 show that the MLP algorithm works best for the MGH dataset. However, the SVM algorithm excelled at the ICL dataset's models in Tasks 2 and 3. As recall is the most important evaluation metric for the specific use case, the best score for MGH and YPS datasets is achieved via MLP, and the ICL dataset is achieved using SVM.
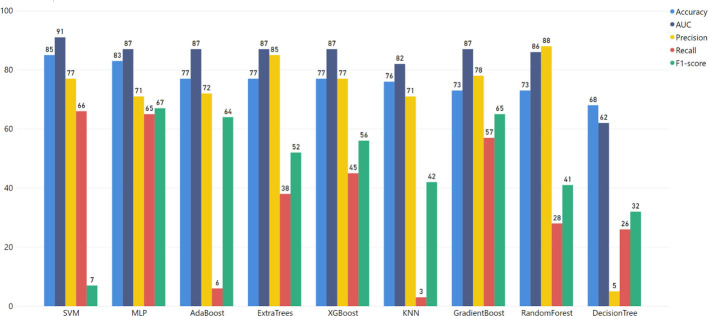
4.4. Testing on an independent dataset
Furthermore, the data from the study of the Mayo Clinic (MC) containing 455 COVID-19 patients [11] was also used to evaluate the MLP model (Supplementary Fig. 13). We followed the methodology of task 1. The results verified MLP as the best algorithm for the MC dataset (Supplementary Table 11, Supplementary Fig. 12). To this end, the ML pipeline mentioned above (Fig. 2) is an optimal procedure to reveal the outstanding algorithm for this type of data.
Supplementary Fig. 13.
The proposed pipeline of our study is evaluated through the MC dataset.
4.5. Explainable AI reveals predictive plasma proteins for COVID-19 severity
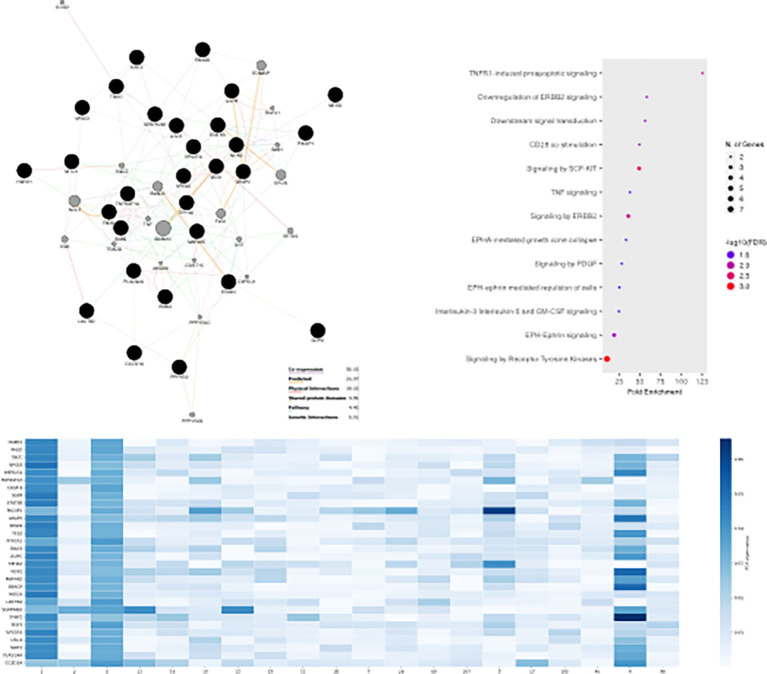
To elucidate the biological significance of plasma proteins representing top-30 loadings of the various ML-approaches (Supplementary Figs. 8, 9, 10), we inferred the PPI network connecting the most predictive proteins for each task and dataset along with the most probable pathways from Reactome.
For Task 1, we predominantly detected signaling cellular responses that have been extensively documented in COVID-19 immunopathology (e.g., “Signaling by the B Cell Receptor (BCR)”, “TNFR1-induced NFkappaB signaling pathway”, “Toll Like Receptor 10 (TLR10)”, “Myd88 dependent cascade” etc.) (Fig. 8 ). Uniquely, the MGH-Task 1 loadings (Fig. 8A) associated with the most diverse repertoire of signaling pathways which contained not only aforementioned cellular responses but also PECAM1 interactions, platelet sensitization by LDL, vesicle-mediated transport and signaling via SCF/c-Kit signaling axis [31]. Based on ARCHS4 data mining, the most predictive protein in the MGH study was a promoter of RAS/JUN kinase signaling pathways called CRKL (CRK Like Proto-Oncogene, Adaptor Protein) with metabolic functionalities while in the YPS study IRAK1 (Interleukin 1 Receptor Associated Kinase 1), a potentiator of IL1R downstream signaling, was the most predictive protein. Furthermore, on the ICL study, the most predictive protein was NEMO/IKBKG which is a critical mediator of the Nf-KB pathway.
Fig. 8.
MLP predicts COVID-19 severity using proteins regulating immune-inflammatory and developmental pathways in Task 1. GeneMania protein-protein interaction networks (PPI), pathway enrichment dotplots and heatmaps with protein-nodes for the most predictive plasma proteins (top-30) as features of MLP (Task 1) considering the MGH (A), YPS (B) and ICL (C) datasets respectively.
For Task 2 (Supplementary Figs. 8, 9), the various ML iterations were accompanied by the enrichment of TLR signaling pathways, IFN signaling and TNF signaling. The most predictive proteins in this task were IRAK1, AXIN1 (Axis Inhibition Protein 1) which impedes on the Wnt pathway and SRPK2 (Serine/Arginine-Rich Protein-Specific Kinase 2) which is a multi-faceted kinase involved in neurotransmitter signaling in the Central Nervous System (CNS).
Lastly, regarding the evaluation dataset of MC, the most predictive proteins correlated with TNF signaling, SCF/c-Kit signaling, PDGF signaling and IL-3/IL-5/GMCSF signaling, resembling the pathway landscape of MGH dataset. The most predictive protein in the MC study was HARS1 (Histidine–TRNA Ligase, Cytoplasmic) which is an aminoacyl-tRNA synthetase (Supplementary Fig. 10).
5. Discussion
A bevy of high-throughput biological studies with clinical annotations has been at the forefront of biomarker discovery for COVID-19 [32]. To this end, AI could be used to provide early hints or useful insights regarding the disease progression and the impact of various factors, including the identification of potential causal factors. Albeit a multitude of different AI approaches are being applied in the aforementioned datasets for COVID-19 (e.g., Random Forest, Logistic regression etc.) [9], there are still several lingering caveats considering predominantly the lack of interpretability and explainability (“black box” challenge) [33]. Acknowledging these hurdles, in the herein work we present a benchmarking pipeline for various ML classifiers based on COVID-19 plasma proteomics (3 datasets based on Olink PEA technology encompassing detailed clinical covariates) engaging an “interpretable” AI approach [34].
Assembling the benchmarking pipeline for ML classifiers was predicated on the analysis of the current literature which highlighted the narrow number of ML algorithms (usually decision tree algorithms) that are already used in proteome studies. Furthermore, most presented scientific studies did not usually select XAI models to spot biomarkers but other feature selection methods e.g. Mean decrease in Gini [35]. Finally, most studies analyze small patient cohorts and plasma proteins while their findings lack external validation with independent datasets.
The benchmarking pipeline consists of 3 Tasks that helped to test several algorithms and to conclude the most optimal ML algorithm for our data. These tasks are designed to avoid results that arise from overfitting. We choose 4 distinct datasets −3 for training (MGH, YPS, ICL dataset for benchmarking pipeline) and one for evaluation (MC)- based on Proximity Extension Assay (PEA) technology from Olink. This technology has been applied in a diverse array of multi-factorial diseases like pestilential infections (e.g., COVID-19), cancer, neurodegenerative conditions (e.g., Alzheimer's) and it has been proven to provide critical information about disease pathobiology due to a large number of proteins analyzed. Compared to Mass Spectroscopy, NPX data are already normalized and more specific [36]. Therefore, there are already 420 studies that are designed with NPX data. Although the relevant literature review highlighted the AUC score as an evaluation metric for the ML models, in our study we used accuracy, F1-score, Recall and Precision to examine thoroughly the performance of our ML models. Finally, we also included two “interpretation” steps. The first step leveraged SHAP values to point out the top 20 most informative PCA embeddings (Supplementary Figs. 6, 7) and the second ranked the magnitude of protein vectors that these top 20 embeddings consist of. (Supplementary Figs. 8, 9, 10) Recent benchmarking ML studies on biological data tend to execute only the procedure that we followed in Task 1 [37], [38].
The benchmarking pipeline highlighted MLP as the superior model for our experiment based on the Recall score that this algorithm succeeds. As our study focus on the best predictive model for severe COVID-19 patients, recall in the evaluation metric the first compare our models on. Moreover, the SVM model also outperformed the ICL models of Task 2 and Task 3. Even in the independent validation dataset, MLP outperforms the rest of the models.
Concerning the biological interpretation of the results, all models contained, as top-loadings, plasma proteins that pertained to critical aspects of COVID-19 pathobiology like aberrant activation of B cells, cytokine storm, TNF signaling, TLR signaling [35], [39]. Interestingly, in Task 1 for the MGH dataset and in the validation MC study, more diverse pathways were retrieved (Fig. 8, Supplementary Fig. 10), like perturbations with cellular adhesion and endothelial damage, exosomal communication among cells, platelet involvement and deregulation of key cytokine regulate adaptive and innate responses as well as stem cell differentiation i.e. SCF/c-Kit signaling [31], [40], [41], [42]. Since not all of these pathways have been explored in-depth in COVID-19 immunopathology, these findings advocate for the use of explainable AI to unravel nascent biological information from voluminous -omic datasets with potential translational significance [43].
In terms of limitations, it should be highlighted that the used datasets focus on early variations of the SARS-COV-2 virus and do not take into account the Delta or Omicron variation which has significantly varied clinical outcomes [44]. Moreover, it should be noted that the presented ML pipeline (as also the vast majority of papers published in the field) has not been clinically validated [45]. Furthermore, we have only used the SHAP values methodology which merely ranked plasma proteins based on their contributions on COVID-19 patient classification from the MLP model. It should be pointed out that SHAP values should be used for the “interpretation” of ML models where the input values satisfy specific requirements (e.g., statistical independence) which are not necessarily true in our case as the input features of the presented ML models cannot be verified as independent. Still, we argue that SHAP values could be used to provide qualitative insights to identify potential input values which heavily impact the outcome/prediction of the respective algorithms. Albeit pathway enrichment showed the biological relevance of the top-ranked proteins, more complex relations among plasma proteomics like non-linear dependencies (which could lead to new biological insights) remained largely elusive. To address this challenge, a potential way forward could be to research a wider array of ML/DL models (e.g., Bayesian Probabilistic Neural Network) along with Bayesian mathematics and combinations of various interpretable ML methodologies (e.g. Individual Conditional Expectation, Partial Dependence Plot etc.).
Overall, in the herein work, we provide a benchmarking pipeline that could help the selection of appropriate ML tools for studying biological “Big Data” from -omic studies. Our approach employs distinct ML metrics to help avoiding potential overfitting and an interpretability component to assist with the biological explainability of ML models. To the best of our knowledge, we are the first to design such a pipeline for Olink PEA COVID-19 plasma proteomics, hence highlighting known and more obscure -from the literature- proteins and pathways relevant to COVID-19 immunopathology that merit further in vitro and clinical investigation. Conclusively, we posit that this type of ML benchmark studies can aid in the design of ML models closer to the biological “ground truth”, hence increasing the possibilities of discovering novel biomarkers and “druggable” targets.
The following are the supplementary data related to this article.
Common proteins among the three selected datasets.
Supplementary Fig. 2.
Task 1 performance from GridSearch hyperparameter tuning A. MGH dataset B. YPS dataset C. ICL dataset.
Supplementary Fig. 3.
Task 2 performance from GridSearch hyperparameter tuning A. The model is trained in MGH dataset and test in ICL dataset B. The model is trained in MGH dataset and test in YPS dataset C. The model is trained in YPS dataset and test in ICL dataset.
Supplementary Fig. 4.
Task 2 performance from GridSearch hyperparameter tuning A. The model is trained in YPS dataset and test in MGH dataset B. The model is trained in ICL dataset and test in YPS dataset C. The model is trained in ICL dataset and test in MGH dataset.
Supplementary Fig. 5.
Task 3 performance from GridSearch hyperparameter tuning.
MLP predicts COVID-19 severity using proteins regulating immune-inflammatory and developmental pathways in Task 2 (Part A). GeneMania protein-protein interaction networks (PPI), pathway enrichment dotplots and heatmaps with protein-nodes for the most predictive plasma proteins (top-30) as features of MLP (Task 1) considering the MGH-ICL (A) and MGH-YPS (B) iterations respectively.
MLP predicts COVID-19 severity using proteins regulating immune-inflammatory and developmental pathways in Task 2 (Part B). GeneMania protein-protein interaction networks (PPI), pathway enrichment dotplots and heatmaps with protein-nodes for the most predictive plasma proteins (top-30) as features of MLP (Task 1) considering the YPS-ICL (A) and YPS-MGH (B) iterations respectively.
Supplementary Fig. 11.
SHAP values of ML models that are presented the contribution of PCA components to the final classification. A. SHAP values of MLP model in evaluation dataset MC.
Supplementary tables
Declaration of competing interest
None declared.
Acknowledgments
Acknowledgements
N/A.
Funding
This work has been funded by INAB|CERTH's institutional funds.
Footnotes
The use of terms “interpretable AI” and “explainable AI” (XAI) for computational approaches aiming to provide “explanations” (e.g., SHAP) is indeed debatable. It is widely acknowledged that these approaches can mainly be used by data scientists rather than clinical professionals.
References
- 1.World Health Organization . May 2022. COVID-19 Weekly Epidemiological Update. [Online]. Available: who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports. [Google Scholar]
- 2.Yu X., Hartana C., Srivastava A., Fergie J. Immunity to SARS-CoV-2: lessons learned. Front. Immunol. 2019;1 doi: 10.3389/fimmu.2021.654165. www.frontiersin.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dias-Audibert F.L., et al. Combining machine learning and metabolomics to identify weight gain biomarkers. Front Bioeng Biotechnol. Jan. 2020;8(6) doi: 10.3389/FBIOE.2020.00006/FULL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang C.H., Lin C.H., Lane H.Y. Machine learning and novel biomarkers for the diagnosis of Alzheimer’s disease. Int J Mol Sci. Mar. 2021;22(5):1–12. doi: 10.3390/IJMS22052761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer Y., et al. Identifying early pulmonary arterial hypertension biomarkers in systemic sclerosis: machine learning on proteomics from the DETECT cohort. Eur Respir J. Jun. 2021;57(6) doi: 10.1183/13993003.02591-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adadi A., Berrada M. Peeking inside the black-box: a survey on explainable artificial intelligence (XAI) IEEE Access. Sep. 2018;6:52138–52160. doi: 10.1109/ACCESS.2018.2870052. [DOI] [Google Scholar]
- 7.Sidak D., Schwarzerová J., Weckwerth W., Waldherr S. Interpretable machine learning methods for predictions in systems biology from omics data. Front Mol Biosci. Oct. 2022;9 doi: 10.3389/fmolb.2022.926623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gisby J., et al. Longitudinal proteomic profiling of dialysis patients with covid-19 reveals markers of severity and predictors of death. Elife. Mar. 2021;10 doi: 10.7554/ELIFE.64827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beltrami A.P., et al. Combining deep phenotyping of serum proteomics and clinical data via machine learning for COVID-19 biomarker discovery. Int. J. Mol. Sci. 2022;23(16):9161. doi: 10.3390/IJMS23169161. Vol. 23, Page 9161. Aug. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yaşar Ş., Çolak C., Yoloğlu S. Artificial intelligence-based prediction of Covid-19 severity on the results of protein profiling. Comput Methods Programs Biomed. Apr. 2021;202 doi: 10.1016/J.CMPB.2021.105996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byeon S.K., et al. Development of a multiomics model for identification of predictive biomarkers for COVID-19 severity: a retrospective cohort study. Lancet Digit Health. Sep. 2022;4(9):e632–e645. doi: 10.1016/S2589-7500(22)00112-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filbin M.R., et al. Longitudinal proteomic analysis of severe COVID-19 reveals survival-associated signatures, tissue-specific cell death, and cell-cell interactions. Cell Rep Med. 2021;2(5):pp. doi: 10.1016/j.xcrm.2021.100287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krishnan S., et al. Metabolic perturbation associated with COVID-19 disease severity and SARS-CoV-2 replication. Mol Cell Proteomics. Jan. 2021;20 doi: 10.1016/J.MCPRO.2021.100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moons K.G.M., et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): explanation and elaboration. Jan. 2015;162(1):W1–W73. doi: 10.7326/M14-0698. [DOI] [PubMed] [Google Scholar]
- 15.Filbin M.R., et al. bioRxiv. 2020. Plasma proteomics reveals tissue-specific cell death and mediators of cell-cell interactions in severe COVID-19 patients. [DOI] [Google Scholar]
- 16.Gisby J., et al. Longitudinal proteomic profiling of dialysis patients with covid-19 reveals markers of severity and predictors of death. Elife. 2021;10:1–30. doi: 10.7554/eLife.64827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su Y., et al. Multi-Omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell. Dec. 2020;183(6) doi: 10.1016/j.cell.2020.10.037. [Online]. Available. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.WHO . 2021. COVID-19 Clinical Management: Living Guidance.https://apps.who.int/iris/bitstream/handle/10665/338871/WHO-2019-nCoV-clinical-web_annex-2021.1-eng.pdf [Online]. Available. [Google Scholar]
- 19.Cheng Y., et al. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. May 2020;97(5):829–838. doi: 10.1016/J.KINT.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.FABIANPEDREGOSA F.Pedregosa, et al. Scikit-learn: machine learning in Python Gaël Varoquaux Bertrand Thirion Vincent Dubourg Alexandre Passos PEDREGOSA, VAROQUAUX, GRAMFORT ET AL. Matthieu Perrot. J. Mach. Learn. Res. 2011;12:2825–2830. http://scikit-learn.sourceforge.net [Online]. Available. [Google Scholar]
- 21.World Health Organization WHO R&D blueprint novel coronavirus COVID-19 therapeutic trial synopsis. World Heal. Organ. 2020;(February 18, 2020):1–9. https://cdn.who.int/media/docs/default-source/blue-print/covid-19-therapeutic-trial-synopsis.pdf [Online]. Available. [Google Scholar]
- 22.Tsai C.F., Sung Y.T. Ensemble feature selection in high dimension, low sample size datasets: parallel and serial combination approaches. KnowlBased Syst. Sep. 2020;203 doi: 10.1016/j.knosys.2020.106097. [DOI] [Google Scholar]
- 23.Vijayan V., Ravikumar A. Study of data mining algorithms for prediction and diagnosis of diabetes mellitus. Int. J. Comput. Appl. 2014;95(17):12–16. https://www.academia.edu/25378193/Study_of_Data_Mining_Algorithms_for_Prediction_and_Diagnosis_of_Diabetes_Mellitus [Online]. Available. [Google Scholar]
- 24.Che D., Liu Q., Rasheed K., Tao X. Decision tree and ensemble learning algorithms with their applications in bioinformatics. Adv Exp Med Biol. 2011;696:191–199. doi: 10.1007/978-1-4419-7046-6_19/COVER. [DOI] [PubMed] [Google Scholar]
- 25.Breiman L. Random forests. Mach. Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. 451. Oct. 2001. [DOI] [Google Scholar]
- 26.Al M., Hasan M., Ahmad S., Khademul M., Molla I. Protein subcellular localization prediction using multiple kernel learning based support vector machine †. Mol Biosyst. 2017;13:785. doi: 10.1039/c6mb00860g. [DOI] [PubMed] [Google Scholar]
- 27.Geurts P., Ernst D., Wehenkel L. Extremely randomized trees. Mach. Learn. 2006;63(1):3–42. doi: 10.1007/S10994-006-6226-1. 631. Mar. 2006. [DOI] [Google Scholar]
- 28.Ma B., Meng F., Yan G., Yan H., Chai B., Song F. Diagnostic classification of cancers using extreme gradient boosting algorithm and multi-omics data. Comput Biol Med. Jun. 2020;121 doi: 10.1016/J.COMPBIOMED.2020.103761. [DOI] [PubMed] [Google Scholar]
- 29.Svozil D., Kvasnička V., Pospíchal J. Introduction to multi-layer feed-forward neural networks. Chemom Intel Lab Syst. Nov. 1997;39(1):43–62. doi: 10.1016/S0169-7439(97)00061-0. [DOI] [Google Scholar]
- 30.Franz M., et al. GeneMANIA update 2018. Nucleic Acids Res. Jul. 2018;46(W1):W60–W64. doi: 10.1093/NAR/GKY311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L., et al. Serum levels of soluble platelet endothelial cell adhesion molecule 1 in COVID-19 patients are associated with disease severity. J Infect Dis. Jan. 2021;223(1):178–179. doi: 10.1093/INFDIS/JIAA642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papadopoulou G., Manoloudi E., Repousi N., Skoura L., Hurst T., Karamitros T. Molecular and clinical prognostic biomarkers of COVID-19 severity and persistence. Pathogens. Mar. 2022;11(3):311. doi: 10.3390/PATHOGENS11030311/S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bjerring J.C., Busch J. Artificial intelligence and patient-centered decision-making. Philos. Technol. 2020;34(2):349–371. doi: 10.1007/S13347-019-00391-6. 342. Jan. 2020. [DOI] [Google Scholar]
- 34.Ennab M., McHeick H. Designing an interpretability-based model to explain the artificial intelligence algorithms in healthcare. Diagnostics. 2022;12(7) doi: 10.3390/diagnostics12071557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Filbin M.R., et al. Longitudinal proteomic analysis of plasma from patients with severe COVID-19 reveal patient survival-associated signatures, tissue-specific cell death, and cell-cell interactions. Cell Rep Med. 2021 doi: 10.1016/j.xcrm.2021.100287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.“PEA-a high-multiplex immunoassay technology with qPCR or NGS readout.” n.d. https://www.olink.com/content/uploads/2021/09/olink-white-paper-pea-a-high-multiplex-immunoassay-technology-with-qpcr-or-ngs-readout-v1.0.pdf.
- 37.Cao X., Xing L., Majd E., He H., Gu J., Zhang X. A systematic evaluation of supervised machine learning algorithms for cell phenotype classification using single-cell RNA sequencing data. Front Genet. Feb. 2022:168. doi: 10.3389/FGENE.2022.836798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azodi C.B., Bolger E., McCarren A., Roantree M., de los Campos G., Shiu S.-H. Benchmarking parametric and machine learning models for genomic prediction of complex traits. G3 Genes|Genomes|Genetics. Nov. 2019;9(11):3691. doi: 10.1534/G3.119.400498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reyes M., et al. Plasma from patients with bacterial sepsis or severe COVID-19 induces suppressive myeloid cell production from hematopoietic progenitors in vitro. Sci Transl Med. 2021;13(598):pp. doi: 10.1126/scitranslmed.abe9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van de Veerdonk F., et al. 2020. A Systems Approach to Inflammation Identifies Therapeutic Targets in SARS-CoV-2 Infection. [DOI] [Google Scholar]
- 41.Borowiec B.M., Volponi A.A., Mozdziak P., Kempisty B., Dyszkiewicz-Konwińska M. Small extracellular vesicles and COVID19—using the ‘Trojan horse’ to tackle the Giant. Cells. Dec. 2021;10(12) doi: 10.3390/CELLS10123383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan W.Y.T., Young B.E., Lye D.C., Chew D.E.K., Dalan R. Statin use is associated with lower disease severity in COVID-19 infection. Sci Rep. Dec. 2020;10(1) doi: 10.1038/S41598-020-74492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anguita-Ruiz A., Segura-Delgado A., Alcalá R., Aguilera C.M., Alcalá-Fdez J. EXplainable Artificial Intelligence (XAI) for the identification of biologically relevant gene expression patterns in longitudinal human studies, insights from obesity research. 2020;16(4) doi: 10.1371/journal.pcbi.1007792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lauring A.S., et al. Clinical severity of, and effectiveness of mRNA vaccines against, covid-19 from omicron, delta, and alpha SARS-CoV-2 variants in the United States: prospective observational study. BMJ. Mar. 2022;376 doi: 10.1136/BMJ-2021-069761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abdulkareem M., Petersen S.E. The promise of AI in detection, diagnosis, and epidemiology for combating COVID-19: beyond the hype. Front Artif Intell. May 2021:53. doi: 10.3389/FRAI.2021.652669. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Common proteins among the three selected datasets.
MLP predicts COVID-19 severity using proteins regulating immune-inflammatory and developmental pathways in Task 2 (Part A). GeneMania protein-protein interaction networks (PPI), pathway enrichment dotplots and heatmaps with protein-nodes for the most predictive plasma proteins (top-30) as features of MLP (Task 1) considering the MGH-ICL (A) and MGH-YPS (B) iterations respectively.
MLP predicts COVID-19 severity using proteins regulating immune-inflammatory and developmental pathways in Task 2 (Part B). GeneMania protein-protein interaction networks (PPI), pathway enrichment dotplots and heatmaps with protein-nodes for the most predictive plasma proteins (top-30) as features of MLP (Task 1) considering the YPS-ICL (A) and YPS-MGH (B) iterations respectively.
Supplementary tables