FIGURE 5.
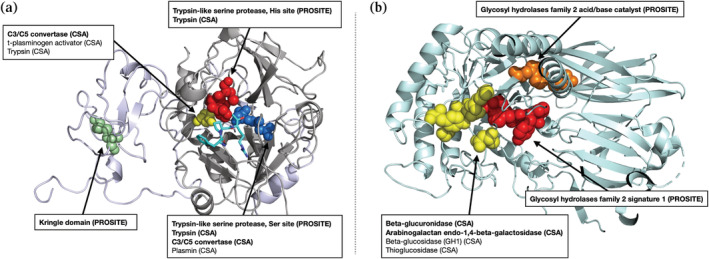
Results of functional annotation tool applied to (a) meizothrombin (PDB ID 1A0H) and (b) beta‐glucuronidase (PDB ID 3HN3), both at p < . No member of the beta‐glucuronidase family is in the training set (maximum sequence identity 2.8%). Functional residues identified by our method are shown as spheres, with colors corresponding to the functional site. Hits labeled in bold are also significant at a more stringent cutoff (p < ). All hits represent either the correct function or those of very closely related proteins, showing that COLLAPSE is effective for annotation of proteins whether or not similar proteins are present in the training set
