Figure 1.
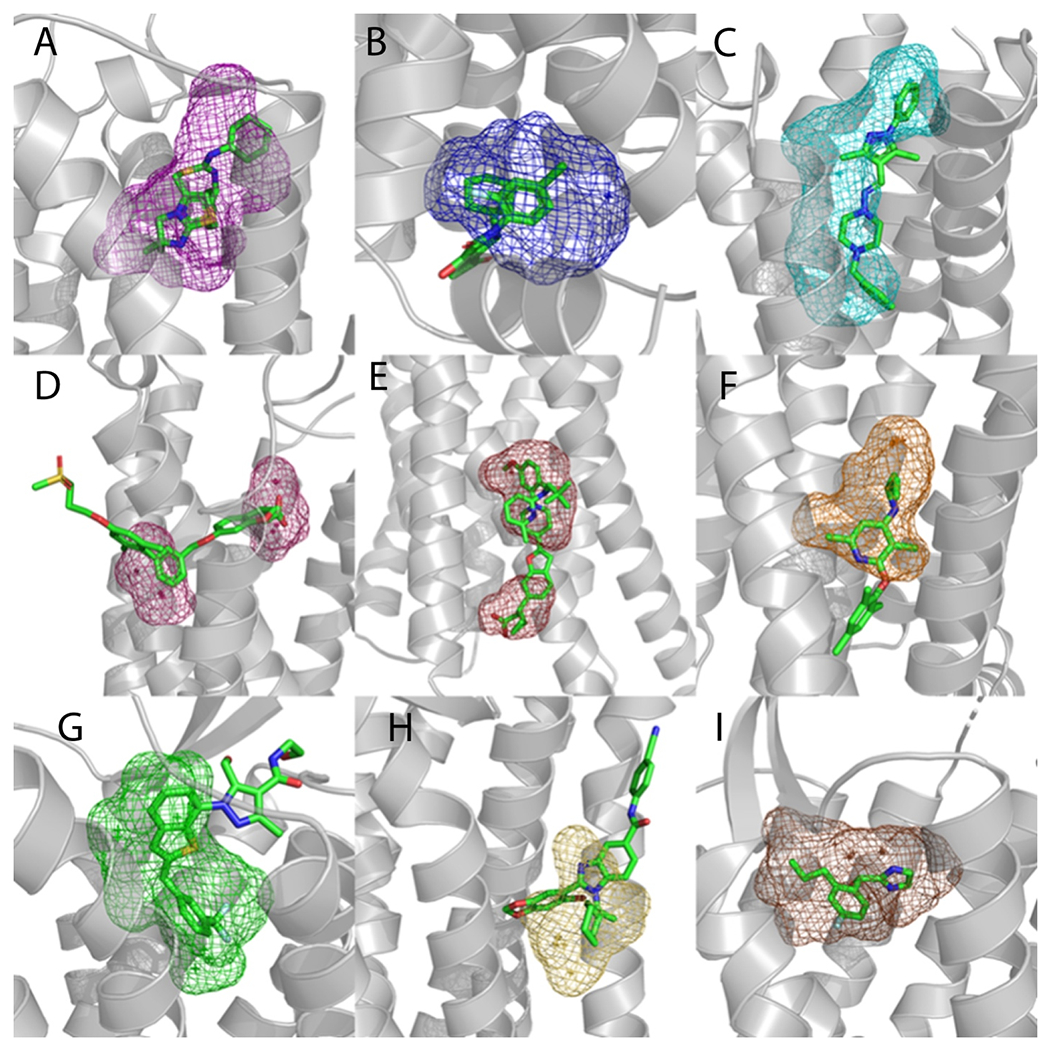
Examples of FTMap site prediction (mesh) in proteins (gray) without co-crystalized allosteric ligands. Binding site predictions were determined by selecting FTMap probe atoms within 3 Å of an allosteric ligand (green sticks) placed by structural alignment. A. Predicted binding pocket in the A2A protein (PDB 3REY) overlayed with the allosteric ligand IT1t from the CXCR4 protein (PDB 3ODU). B. Predicted binding pocket in the GPR52 protein (PDB 6LI1) overlayed with the allosteric ligand C17 from the CCR2 protein (PDB 5T1A). C. Predicted binding pocket in the DRD2 protein (PDB 6CM4) overlayed with the allosteric ligand SANT-1 from the SMO protein (PDB 4N4W). D. Predicted binding pocket in the LPAR1 protein (PDB 4Z34) overlayed with the allosteric ligand TAK-875 from the FFAR1 protein (PDB 4PHU). E. Predicted binding pocket in the P2Y12 protein (PDB 4PXZ) overlayed with the allosteric ligand Compound 1 from the FFAR1 protein (PDB 5KW2). F. Predicted binding pocket in the GLR protein (PDB 5YQZ) overlayed with the allosteric ligand CP-376395 from the CRFR1 protein (PDB 4K5Y). G. Predicted binding pocket in the AGTR1 protein (PDB 4YAY) overlayed with the allosteric ligand C17 from the FFAR1 GPR52 (PDB 6LI0). H. Predicted binding pocket in the PE2R3 protein (PDB 6AK3) overlayed with the allosteric ligand AZ3451 from the PAR2 protein (PDB 5NDZ). I. Predicted binding pocket in the CXCR4 protein (PDB 3OE8) overlayed with the allosteric ligand AZ8838 from the PAR2 protein (PDB 5NDD).
