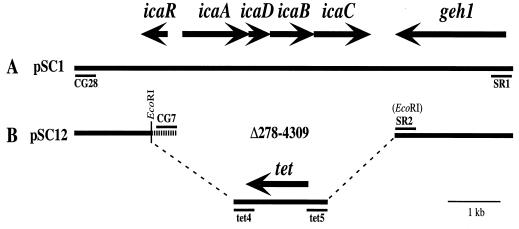
FIG. 1.
Schematic representation of cloning strategy used to construct the ica deletion-replacement mutant of S. epidermidis. A similar strategy was used to construct the ica deletion-replacement mutant of S. aureus, described in reference 7. (A) Plasmid pSC1. Sequences including and surrounding the ica locus are included in database entry U43366. Sequences including and surrounding the geh1 gene are included in database entry AF053006. PCR primers used to amplify cloned DNA are indicated (CG-28 and SR-1). (B) Plasmid pSC12, the deletion-replacement construct used for homologous recombination. Inverse PCR primers used to delete the ica locus (CG-7 and SR-2) and amplify the tetracycline resistance cassette (tet4 and tet5) are indicated. EcoRI restriction recognition sites contained within the sequences of primers SR-2, tet-4, and tet-5 and the EcoRI restriction site in the middle of icaR were used to insert the tetracycline resistance cassette. Deleted sequence numbers refer to database sequence U43366.