Fig. 1 |. iSPI is a multi-purpose, regenerable, proteome-scale human phosphopeptide resource for phosphoproteoimcs.
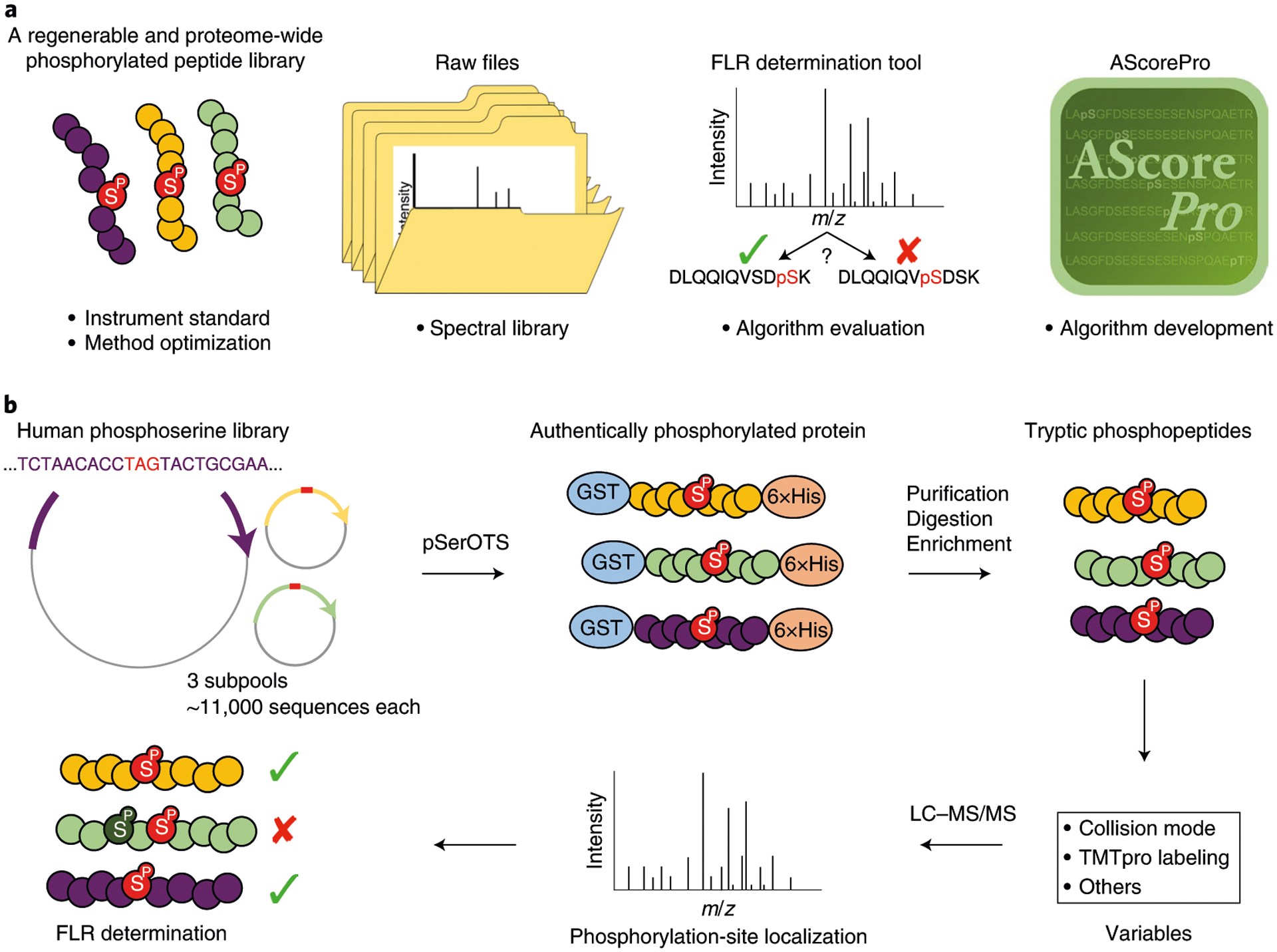
a, iSPI generates authentically phosphorylated peptides derived from the human phosphoserine proteome that can be used as standards and for instrumental method optimization. The raw data files can be used to generate ground-truth phosphopeptide spectral libraries. The FLR determination tool enables researchers to investigate their own data and data processing workflows. AScorePro is an improved version of AScore, with improved phosphorylation-site localization. b, Workflow for determining FLR using iSPI. For this report, 3 library subpools of ~11,000 phosphorylated peptides were expressed using the Phosphoserine Orthogonal Translation System (pSerOTS), which, after purification, digestion, and enrichment, generated 3 pools of peptides with known phosphorylation positions. Note that, for clarity, each subpool never contained the same phosphopeptide phosphorylated on two sites (no positional isomers). Tryptic phosphorylated peptides were analyzed under various collision modes with and without TMTpro labeling, and search-engine outputs were compared with the known positions to determine FLR. LC–MS/MS, liquid chromatography–tandem mass spectrometry.
