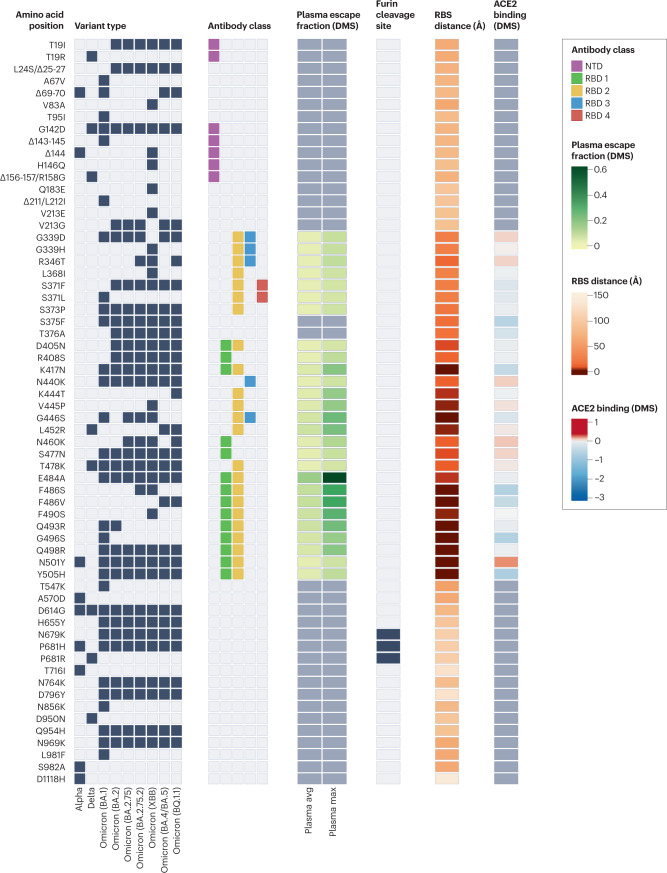
Fig. 1. Properties of amino acid substitutions or deletions in selected SARS-CoV-2 variants of concern.
Black boxes denote the presence of each mutation in the variant of concern. Epitope residues are coloured to indicate the amino-terminal domain (NTD) supersite187 or the receptor-binding domain (RBD) class188. For RBD residues, the results of deep mutational scanning (DMS) studies show the escape fraction (that is, a quantitative measure of the extent to which a mutation reduced polyclonal antibody binding) for each mutant averaged across plasma (‘plasma avg’) and for the most sensitive plasma (‘plasma max’)189, illustrating consistency or variation in the effect of a mutation depending on differences in the antibody repertoire of individuals. Mutations in the furin cleavage site are highlighted. Orange shading indicates the distance to angiotensin-converting enzyme 2 (ACE2)-contacting residues that form the receptor-binding site (RBS). Note that the RBS is defined as residues with an atom <4 Å from an ACE2 atom in the structure of the RBD bound to ACE2 (RCSB Protein Data Bank ID 6M0J190). Finally, ACE2-binding scores representing the binding constant (Δlog10 KD) relative to the wild-type reference amino acid from DMS experiments are shown in shades of red or blue26.