Fig. 2. Furin cleavage site and variant success.
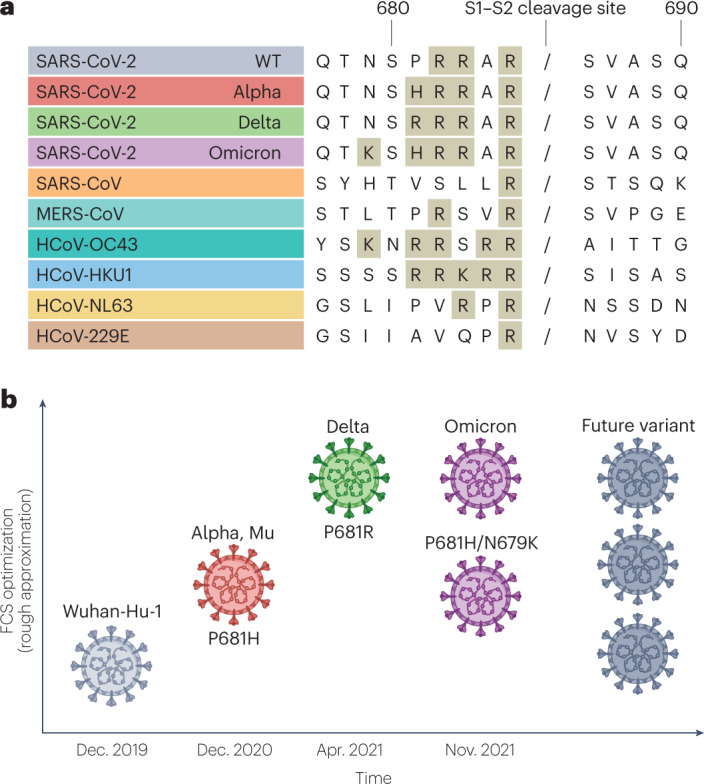
a, Comparison of S1–S2 cleavage site sequences in wild-type (WT) severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and Alpha, Delta and Omicron variants of concern compared with other coronaviruses: SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), human coronavirus OC43 (HCoV-OC43), HCoV-HKU1, HCoV-NL63 and HCoV-229E. A slash indicates the putative furin/serine protease cleavage site. Amino acids contributing to monobasic or polybasic cleavages sites are shaded. b, Diagrammatic representation of estimated relative optimization of the S1–S2 furin cleavage site (FCS) for the variants of concern. The mutations affecting FCS function are indicated. Note that non-concordant results have been observed for Omicron, as indicated. The level of FCS optimization for future variants is uncertain. Data on Alpha from refs. 19,24,25, data on Delta from refs. 22,23,69 and data on Omicron from refs. 32,75,76.
