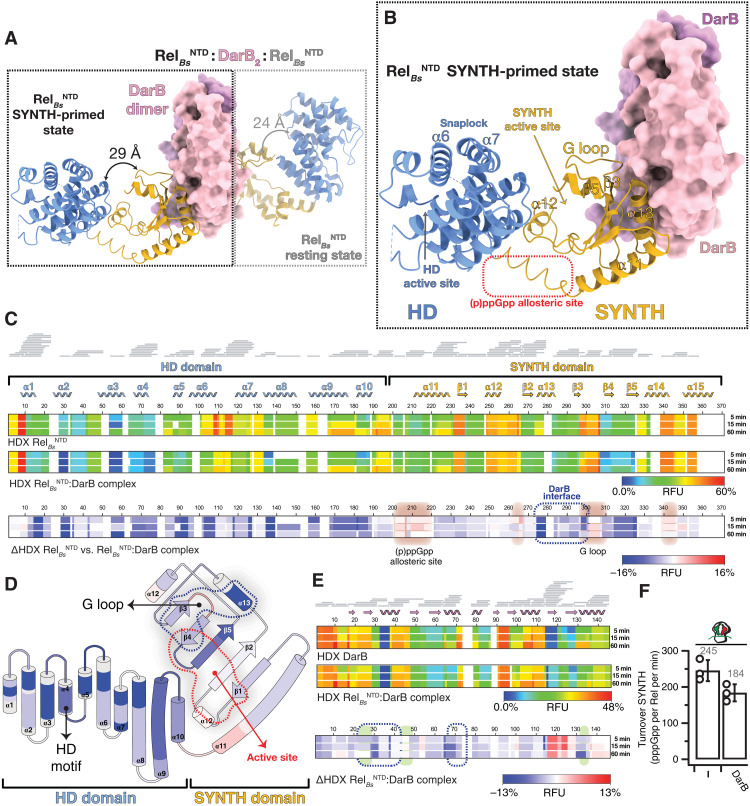
Fig. 2. Structure of the RelBsNTD2:DarB heterotetrameric complex.
(A) Crystal structure of DarB2:RelBsNTD2 heterotetrameric complex with the disc-shaped DarB dimer located at the center of the complex (colored in pink and lilac) and the two RelBsNTD bound at both sides of the DarB dimer. For each RelBsNTD molecule of the complex, the HD domain is colored in light blue, and the SYNTH domain is in yellow. In the nonsymmetrical hetero-complex, the RelBsNTD in the SYNTH-primed state (left, outlined with a black dashed line) is observed in a more open and less structured conformation than the resting RelBsNTD molecule (right, outlined with a light gray dashed line). The relative HD-to-SYNTH distance in each RelBsNTD monomer is indicated. (B) Details of RelBsNTD in the SYNTH-primed state highlighting key structural elements. (C) Heatmaps showing the HDX signal kinetics of RelBsNTD (top) and RelBsNTD as part of the DarB2:RelBsNTD2 complex (center) and the ΔHDX (bottom). Both catalytic domain of RelBs and all the secondary structural elements of the NTD are shown in the figure. Residues involved in the binding interface with DarB are outlined by a dashed blue line, and the regions with increased deuterium uptake, which include the G loop that becomes exposed upon binding to DarB and the alarmone allosteric site, are shaded in red. (D) Topology representation of RelBsNTD colored as a function of the ΔHDX. (E) Heatmaps representing the HDX of DarB (top) and DarB as part of the DarB2:RelBsNTD2 complex (center) and the ΔHDX (bottom). Residues involved in the binding interface with RelBsNTD are indicated by a dashed blue line, and those involved in the binding to c-di-AMP are shaded in green. (F) Effect of DarB on the SYNTH activity of RelBs in the presence or absence of “starved” ribosomes.