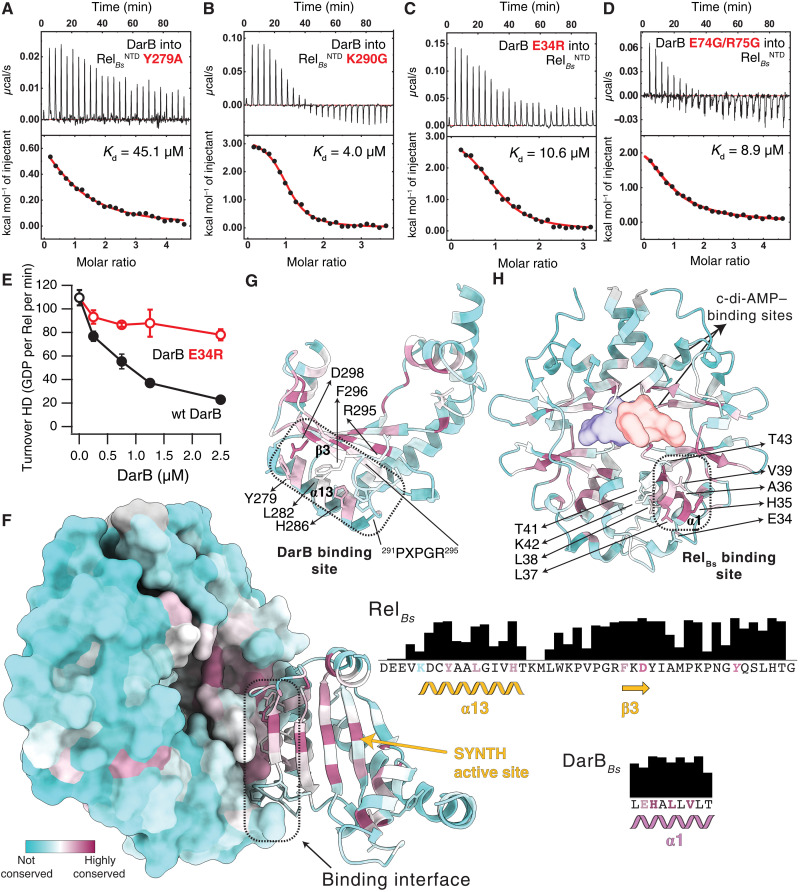
Fig. 4. Biophysical and biochemical interrogation of the RelBs:DarB binding interface.
Effect of the Y279A (A) and K290G (B) substitutions in RelBsNTD to the interaction with DarB, monitored by ITC. Effect of the substitutions to the primary E34R (C) or secondary E74G/R75G (D) DarB interfaces, to the binding to RelBsNTD, monitored by ITC. (E) Effect of an E34R substitution on DarB to the activity of DarB monitored as a function of the hydrolase activity of RelBs. (F) DarB-SYNTH interface color-coded by the conservation score of each amino acid calculated by ConSurf. Residues involved in the primary interface are shown in the conservation bar plots to the right and colored on the basis of their individual conservation profile. The strictly conserved Y of the G loop of Rel is shown in italic. Structural elements of SYNTH (G) and DarB (H) colored as in (F) underline the strong conservation of the binding interface. The contact regions between both proteins are outlined by dashed black lines with the residues directly involved in the primary binding interface and the PXPGR motif highlighted in (G) and (H) and the location of the c-di-AMP shown as a surface in (H).