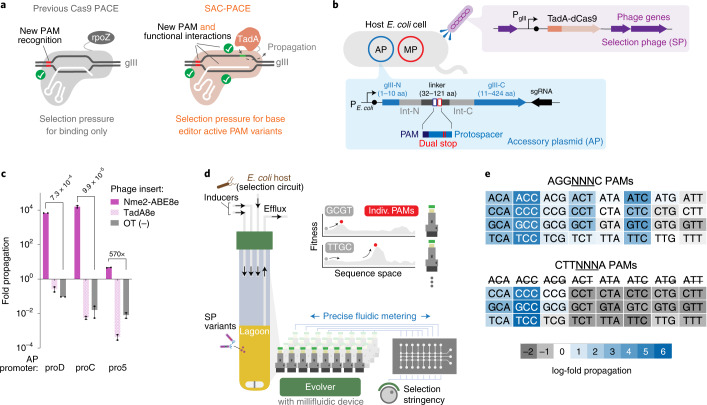
Fig. 1. Development of a function-dependent Cas9 selection and the ePACE platform for automated parallel evolution.
a, Overview of previous Cas9 PACE (left) requiring only PAM binding, compared to the SAC-PACE developed in this study, which requires both PAM binding and subsequent base editing. b, The selection circuit in SAC-PACE. The SP encodes an adenine base editor in place of gIII. In the host cells, an accessory plasmid (AP) contains a cis intein-split gIII, with a linker (31–121 aa) containing stop codons. MP, mutagenesis plasmid 6 (ref. 39). c, Overnight phage propagation assays to test the selection stringency of SAC-PACE with various AP promoter strengths. Mean ± s.e.m. are shown and are representative of n = 2 independent biological replicates. d, Overview of ePACE, enabling parallel lagoon evolution of a Cas9 variant on single PAMs (Supplementary Figs. 1–4). e, Overnight propagation assays of wild-type Nme2-ABE8e on two sets of 32 N3NYN PAMs. Fold-propagation was measured by qPCR and is reflective of the average of two independent biological replicates. The eight CTTAYNA PAMs are excluded as they introduce an additional stop codon in the accessory plasmid, preventing Cas-dependent propagation.