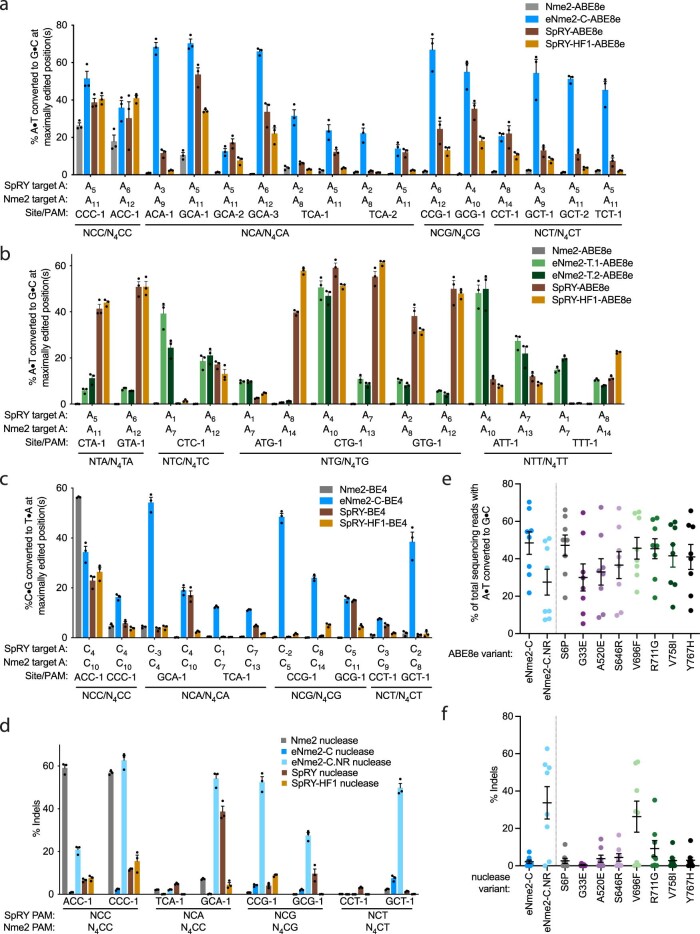
Extended Data Fig. 7. eNme2 variants compared to SpRY and SpRY-HF1 in HEK293T cells.
(a) Adenine base editing activity of eNme2-C-ABE8e compared to SpRY-ABE8e and SpRY-HF1-ABE8e at 14 NCN/N4CN PAM-matched sites in HEK293T cells (pooled data in Fig. 3b). (b) Adenine base editing activity of eNme2-T.1-ABE8e and eNme2-T.2-ABE8e compared to SpRY-ABE8e and SpRY-HF1-ABE8e at eight NTN/N4TN PAM-matched sites in HEK293T cells (pooled data in Fig. 3c). (c) Cytosine base editing activity of eNme2-C-BE4 compared to SpRY-BE4 and SpRY-HF1-BE4 at eight NCN/N4CN PAM-matched sites in HEK293T cells (pooled data in Fig. 3d). (d) Nuclease activity of eNme2-C nuclease and eNme2-C.NR nuclease compared to SpRY nuclease and SpRY-HF1 nuclease at eight NCN/N4CN PAM-matched sites in HEK293T cells (pooled data in Fig. 3e). For (a-d), Mean±SEM is shown and reflects the average activity and standard error of n = 3 independent biological replicates measured at the maximally edited position (if applicable) within each given genomic site. (e) Pooled adenine base editing activity of eNme2-C-ABE8e compared to eNme2-C.NR-ABE8e or adenine base editors generated from reversion mutations at each of the eight RuvC/HNH domain mutations in eNme2-C at eight genomic sites in HEK293T cells. (f) Pooled nuclease activity of eNme2-C nuclease compared to eNme2-C.NR nuclease or nuclease-active variants generated from reversion mutations at each of the eight RuvC/HNH domain mutations in eNme2-C at eight genomic sites in HEK293T cells. For (e) and (f), each point represents the average of n = 3 independent biological replicates measured at the maximally edited position within each given genomic site in HEK293T cells. Mean±SEM is shown and reflects the average activity and standard error of the pooled genomic site averages.