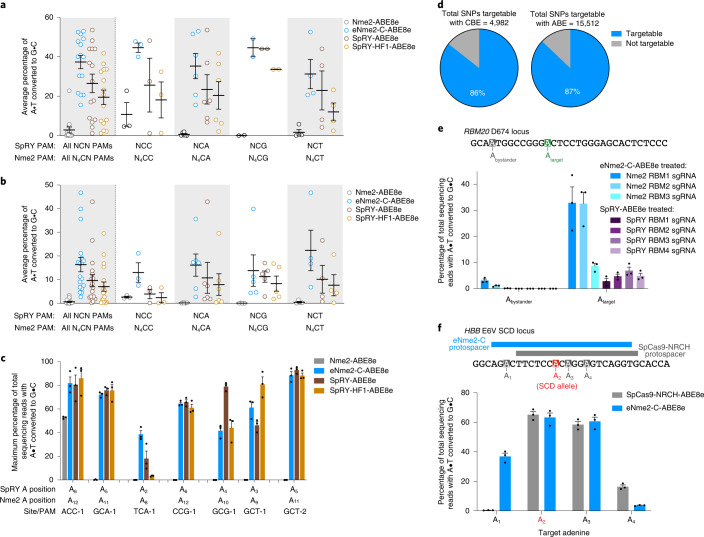
Fig. 4. Generalizability of eNme2-C-ABE8e across different cell types and targets.
a, Summary dot plots showing the activity of eNme2-C-ABE8e compared to SpRY-ABE8e and SpRY-HF1-ABE8e at 15 PAM-matched NCN/N4CN sites in HUH7 cells. Left-most data represent a summary of all 15 sites, and subsequent columns represent a subdivision into specific PAMs. b, Summary dot plots showing the activity of eNme2-C-ABE8e compared to SpRY-ABE8e and SpRY-HF1-ABE8e at 18 PAM-matched NCN/N4CN sites in U2OS cells. Left-most data represent a summary of all 18 sites, and subsequent columns represent a subdivision into specific PAMs. For a and b, each point represents the average editing of n = 3 independent biological replicates measured at the maximally edited position within each given genomic site. Mean ± s.e.m. is shown and reflects the average activity and standard error of the pooled genomic site averages. c, eNme2-C-ABE8e compared to SpRY-ABE8e and SpRY-HF1-ABE8e at eight PAM-matched NCN/N4CN sites in HDFa cells. Bars represent mean ± s.e.m. of n = 3 independent biological replicates, with individual values shown as dots. d, ClinVar identified SNPs that can be targeted with an eNme2-C-ABE8e (top) or eNme2-C-BE4 (bottom). e, Installation of a disease-relevant D674G mutation in the RBM20 gene. Tiled guides were used to install the mutation either with eNme2-C-ABE8e or SpRY-ABE8e (see Supplementary Table 5 for sgRNA sequences). f, Conversion of the sickle-cell disease-causing HBB E6V mutation to the benign E6A (Makassar hemoglobin) allele using either SpCas-NRCH-ABE8e or eNme2-C-ABE8e. Bars represent mean ± s.e.m. of n = 3 independent biological replicates, with individual values shown as dots.