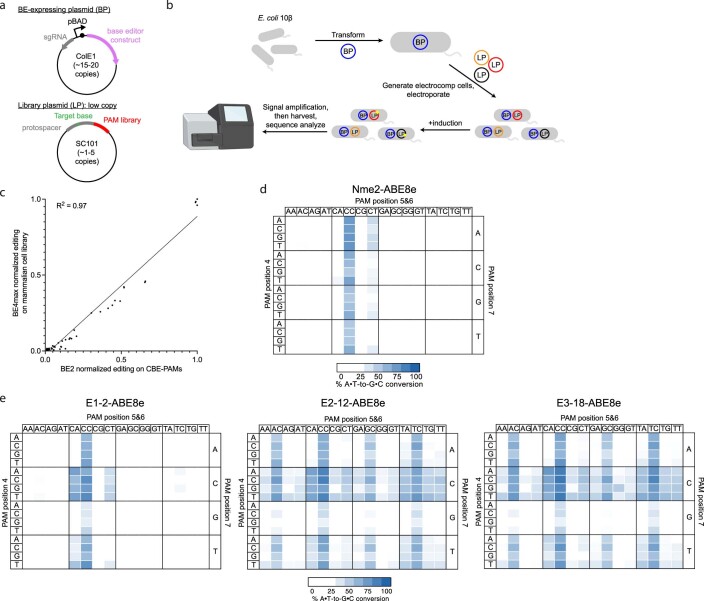
Extended Data Fig. 2. Base editing dependent PAM profiling assay (BE- PPA).
(a) Schematic of BE-PPA constructs. A BE-expressing plasmid (BP) containing the base editor to be evaluated is cloned along with a library plasmid (LP) containing a target protospacer and target base (adenine or cytosine for ABE-PPA or CBE-PPA, respectively) flanked by a library of PAMs of interest. (b) BE- PPA workflow. A cell line containing the BP is first generated, then the LP is electroporated into that cell line before base editor expression is induced. Induced cells are grown for 22–36 hours (with dilution after 24 hours if necessary), before plasmid DNA is harvested and sequenced by high-throughput sequencing. (c) Comparison of the BE-PPA assay against existing mammalian cell base editing PAM profiling9. Each point represents 1 of 64 NNN PAMs, normalized to the activity of the highest PAM for BE2 (rAPOBEC1-dSpCas) along the x- axis in BE-PPA or for BE4max along the y-axis for the previously assessed mammalian library. All points reflect the average normalized activity of n = 2 independent biological replicates. The line reflects a simple ordinary least squares (OLS) regression, with the R-squared value shown. (d, e) Heat maps showing ABE-PPA activity of (d) wild-type Nme2-ABE8e and (e) representative clones from ePACE1-3 on the set of 256 N3NNNN PAMs (PAM positions 1-3 fixed, see Supplementary Table 1). Values are raw % A•T-to-G•C conversion observed for one replicate of each base editor.