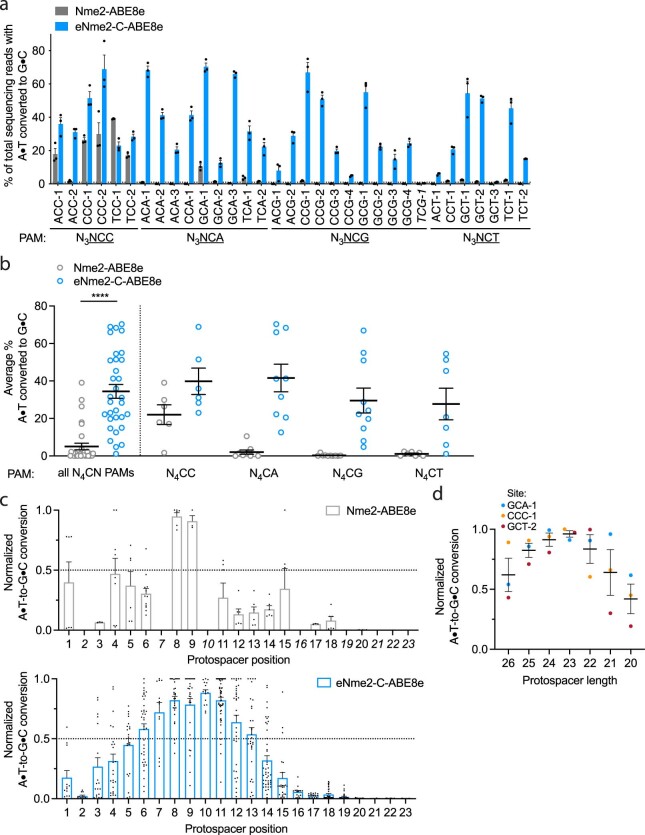
Extended Data Fig. 4. N4CN activity, editing window, and preferred spacer length of eNme2- C-ABE8e.
(a) Adenine base editing activity of eNme2-C-ABE8e at 33 N3NCN PAM-containing sites in HEK293T cells. Mean±SEM is shown and reflects the average activity and standard error of n = 3 replicates at the maximally edited position within each genomic site. The site that exhibited <1% base editing activity (line shown) that was excluded in subsequent analyses is italicized. (b) Pooled adenine base editing activity of eNme2-C- ABE8e from (a). Left: all 32 sites with base editing >1% for eNme2-C-ABE8e; right: sites pooled by PAM position 6 identity. Each point represents the average editing of n = 3 independent biological replicates measured at a given genomic site. Mean±SEM is shown and reflects the average activity and standard error of the pooled genomic site averages. (c) Editing window of Nme2-ABE8e (top) or eNme2- C-ABE8e (bottom) reflective of pooled adenine base editing activity at all 23 protospacer positions (PAM counted as positions 24-29). Each point represents the % A•T-to-G•C conversion observed for an adenine that was present in one of the 32 protospacers, normalized to the highest editing observed within that protospacer. Italicized positions were not present in any protospacers evaluated. Mean±SEM is shown and reflects the average normalized activity and standard error at all observed adenines at that position. (d) Adenine base editing activity of eNme2-C-ABE8e as a function of protospacer length (between 26-20 nt) at three different genomic sites in HEK293T cells. Each point represents the average of n = 3 independent biological replicates observed for a given protospacer length at one genomic site, normalized to the protospacer length with the highest base editing activity for that site. Mean±SEM is shown and reflects the average normalized activity and standard error of the pooled averages at the observed sites. ****, p } 0.0001 by unpaired, two-tailed Student’s t-test.