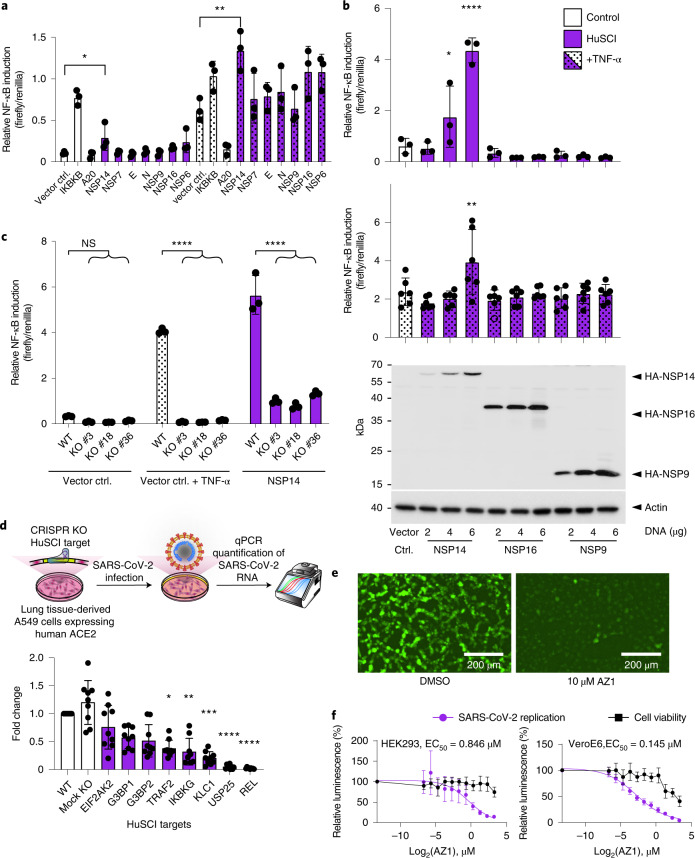
Fig. 4. Validation of pathways and host targets.
a, Relative NF-κB transcriptional reporter activity in unstimulated (left) and TNF-α-stimulated conditions (one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test, P = 0.0395 and P = 0.0047, respectively). Error bars represent standard deviation of the mean, n = 3. b, Relative NF-κB transcriptional reporter activity at different amounts of transfected viral protein-encoding plasmid in unstimulated (top) and TNF-α-stimulated conditions (middle) (one-way ANOVA with Dunnett’s multiple comparisons test: *P = 0.0183, ****P < 0.0001 and **P = 0.0012, respectively). Error bars represent standard deviation of the mean, n = 3 (top) and n = 6 (middle). Representative anti-hemagglutinin (HA) western blot demonstrating levels of tagged viral protein in titration experiments relative to actin beta (ACTB) loading controls (bottom). a and b, Precise P values, biological repeats and n for each test are shown in Extended Data Fig. 4 and Supplementary Table 9. c, Relative NF-κB transcriptional reporter activity under unstimulated (left), TNF-α-stimulated (middle) and NSP14-induced conditions in wild-type (WT) and three independent IKBKG KO clones of HEK293 cells (two-way ANOVA with Dunnett’s multiple comparisons test). Error bars represent standard deviation of the mean, n = 3. Precise P values, biological repeats and n for each test are shown in Extended Data Fig. 4. ctrl., control. d, Schematic of viral replication assay (top) and viral replication in wild-type, mock KO and CRISPR KOs of the indicated HuSCI host targets (bottom) (Kruskal–Wallis with Dunn’s multiple comparisons test, * P = 0.031, ** P = 0.0047, *** P = 0.0003, **** P < 0.0001, respectively). Error bars represent standard deviation of the mean, n = 9. Precise P values, biological repeats and n for each test are shown in Extended Data Fig. 4 and Supplementary Table 10. e, Fluorescence microscopy images showing replication of icSARS-CoV-2-mNeonGreen in infected Vero E6 cells treated with 10 µM AZ1 or solvent (DMSO, dimethylsulfoxide). f, Cell viability and relative replication of icSARS-CoV-2-nanoluciferase in HEK293 cells (left) and Vero E6 cells (right) at different concentrations of AZ1. The EC50 values were calculated with a variable slope model. Error bars represent standard deviation of the mean, n = 8 biological repeats and full analysis in Supplementary Table 11.