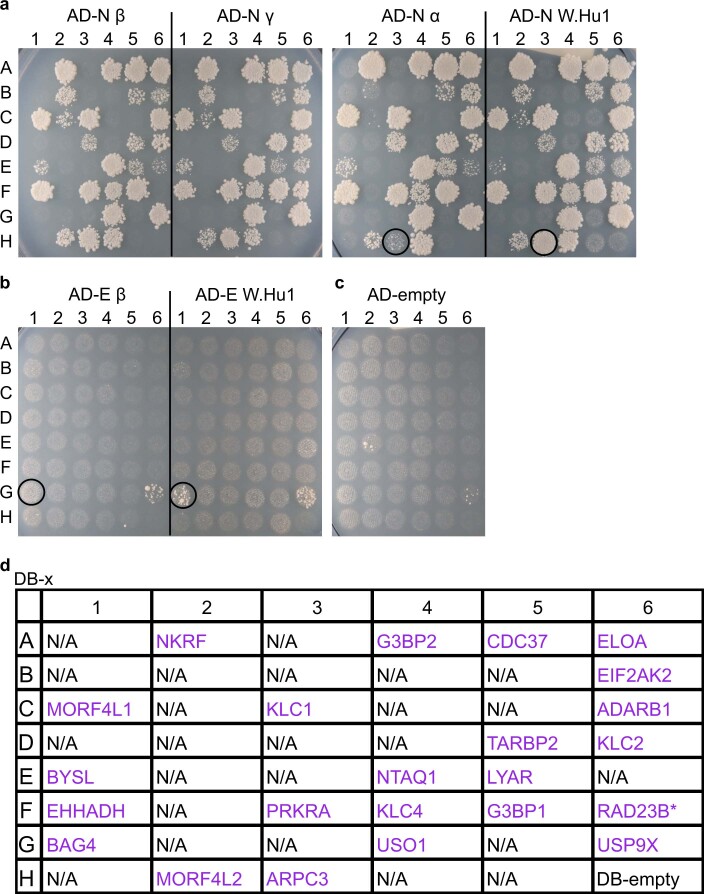
Extended Data Fig. 5. Mutations of SARS-CoV-2 variants affect specific interactions with uSCI host targets.
a, Y2HHIS3 yeast growth on selective plates of HuSCI interaction partners as DB-fusion proteins tested against AD-fusion of the SARS-CoV-2 Nucleocapsid (AD-N) protein (Wuhan-Hu1, original screen) and AD-N containing ‘lineage defining’ amino acid substitutions: D3L and S235F (α-strain), T205I (β-strain), or P80R (γ-strain). Shown is one representative result of 5 repeats. b, Y2HHIS3 yeast growth on selective plates of HuSCI interaction partners as DB-fusion proteins tested against AD-fusion of the SARS-CoV-2 Envelope (AD-E) protein (Wuhan-Hu1, original screen) or AD-E containing ‘lineage defining’ substitution P71L (β-strain). Shown is one representative Y2HHIS3 result on selective media, out of 2 repeats. a - c, Black circles indicate changes in yeast colony growth between human proteins tested against viral variant ORFs or the originally screened Wuhan strain ORFs observed consistently across all repeats. c, AD-empty control plate for a, b indicates lack of autoactivation. d, Layout of DB-fusion HuSCI interactors (purple) tested with AD fusion SARS-CoV-2 proteins or AD-empty control, respectively in a - c. N/A indicates human interactors.