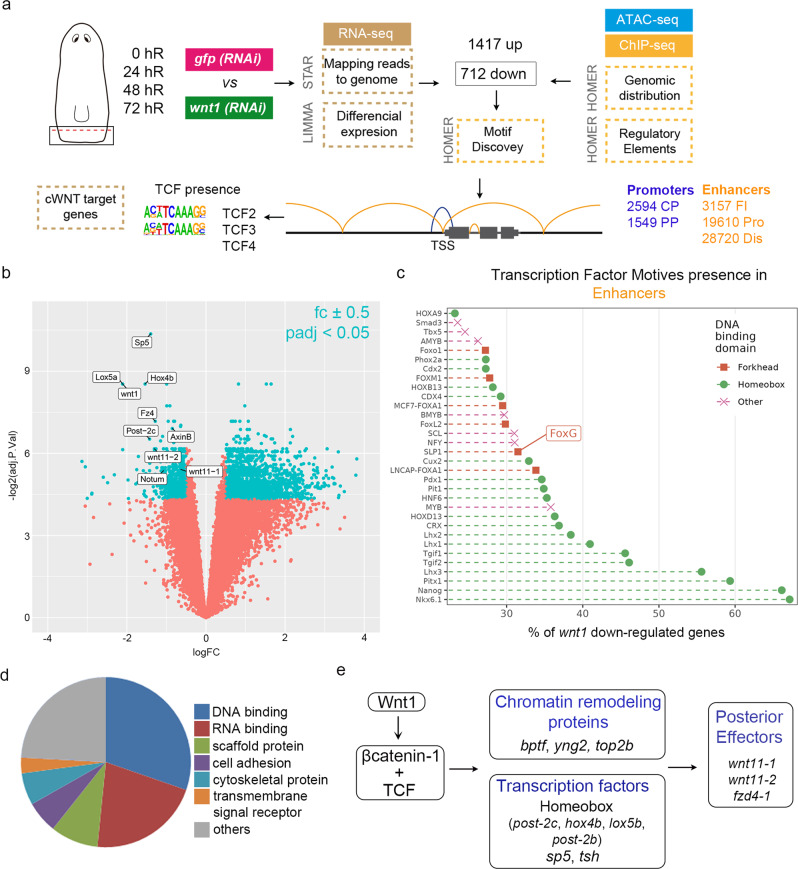
Fig. 2. Homeobox TFs motifs are found enriched in cis-regulatory elements of wnt1 (RNAi) downregulated genes.
a Workflow to identify differentially expressed genes, cis-regulatory elements (CRE) and transcription factors (TFs) associated with wnt1 (RNAi). Next to each workflow the program used is indicated. Motif discovery for TCF binding sites was specifically performed in downregulated wnt1 (RNAi) genes. b Volcano plot of 72 hR shows the (694) down- and (1409) upregulated genes after wnt1 inhibition, which present fold change (fc) ± 0.5 and p-value adjusted (padj) <0.05 (FDR on lima-voom empirical Bayes moderated t-test). Significant genes are colored light blue, and not significant are colored light red. Some significant genes are labeled. Data provided in Supplementary Data 3. c TF motifs presence on the putative enhancer regions of wnt1 (RNAi) downregulated genes, showing a higher representation of Homeobox. Data provided in Supplementary Data 4. d Gene Ontology (molecular function) analysis of the wnt1 (RNAi) downregulated genes containing TCF binding sites (http://pantherdb.org/). Data provided in Supplementary Data 5. e Schematic illustration of the proposed genetic program activated by wnt1 in posterior wounds.