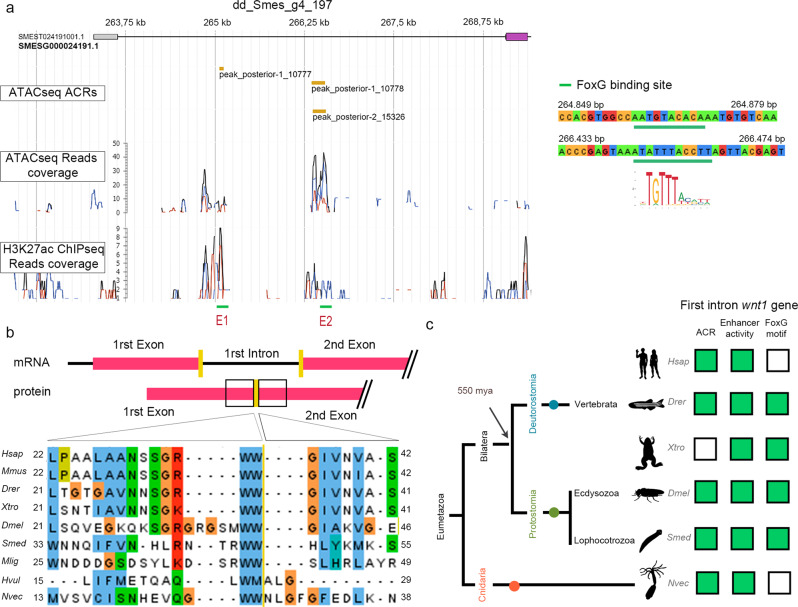
Fig. 3. FoxG could bind to a cis-regulatory element found in wnt1 first intron.
a Schematic illustration of Smed-wnt1 gene locus, indicating exons (violet boxes) linked by introns (lines). Genome Browser screenshot showing the ATAC-seq Accessible Chromatin Regions (ACRs), ATAC-seq and H3K27ac ChIP-seq profiles of the putative enhancer regions. Putative Enhancer 1 (E1) and Enhancer 2 (E2) present a FoxG motif (SLP1) (green line). The ATAC-seq peaks corresponding to E1 and the E2 are indicated. b Alignment of WNT1 amino acid sequences show the conservation of the intron 1 position. Yellow line shows the separation between the first and the second exon. c Schematic summary of accessible chromatin regions (ACRs), enhancer activity and FoxG motif evidence in the first intron of wnt1 genes in different eumetazoan species. Green box indicates evidence and white box indicates no available data. Data is provided in Supplementary Data 7. Species used: Homo sapiens (Hsap), Mus musculus (Mmus), Danio rerio (Drer), Xenopus tropicalis (Xtro), Drosophila melanogaster (Dmel), Schmidtea mediterranea (Smed), Macrostumum ligano (Mlig), Hydra vulgaris (Hvul), and Nematostella vectensis (Nvec). Silhouettes are from https://beta.phylopic.org. Xenopus silhouette is from Sarah Werning and the licence is https://creativecommons.org/licenses/by/3.0/. Schmidthea silhouette is from Noah Schlottman and the license is https://creativecommons.org/licenses/by/3.0/.