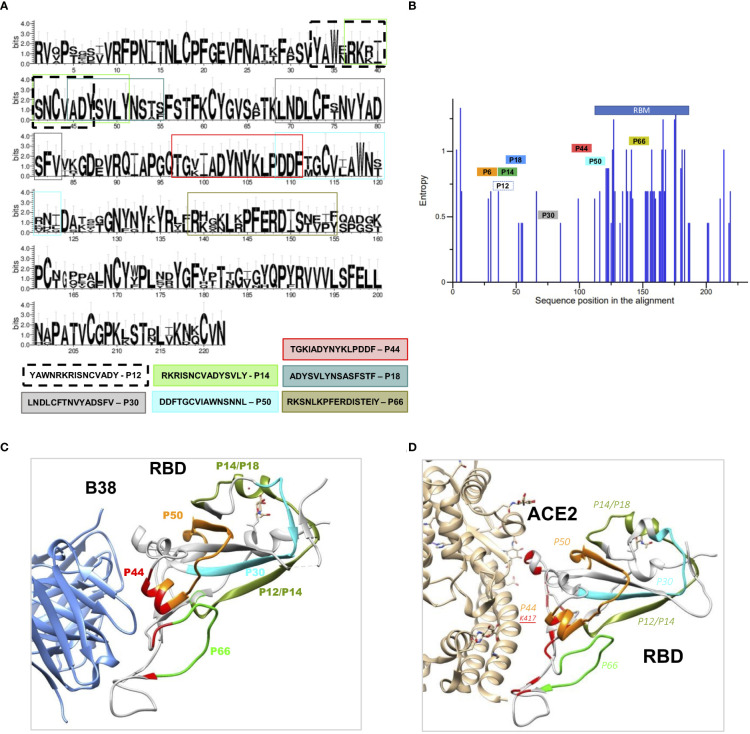
Figure 2.
Structural and conservation evaluation of topmost recognized peptides (A) Conservation analysis of the top recognized peptides among RBD proteins from coronaviruses of the B lineage, SARS-CoV-2, SARS-CoV, SARS-SZ3, Rs3367-bat, CoV-pangolin, and RaTG13-bat. The height of the letters in the logo plot reflects the frequency of the amino acid in the multiple sequence alignment. (B) Entropy of aminoacid residues, top peptides highlighted, of the alignment made at (A, C). RBD binding to the therapeutic neutralizing antibody B38 (pdb: 7BZ5) and with ACE-2 (D) interacting with P44, with contact amino acid residues in red. SARS-CoV-2 RBD (light gray). Colored regions show the most frequently recognized peptides identified among convalescents, namely, orange: P44: peptide 44 (S415-429), silver P6: peptide 6 (S353-367), golden P12, P14 and P18, peptide 12, 14, and 18 (S365-379, S370-384, S378-392), blue P30: peptide 30 (S397-401), violet P50, peptide 50 (S427-441) and green P66, peptide 66 (S459-473).