Figure 1. Recurrent patterns of complex genomic rearrangements (CGRs) in constitutional and cancer genomes.
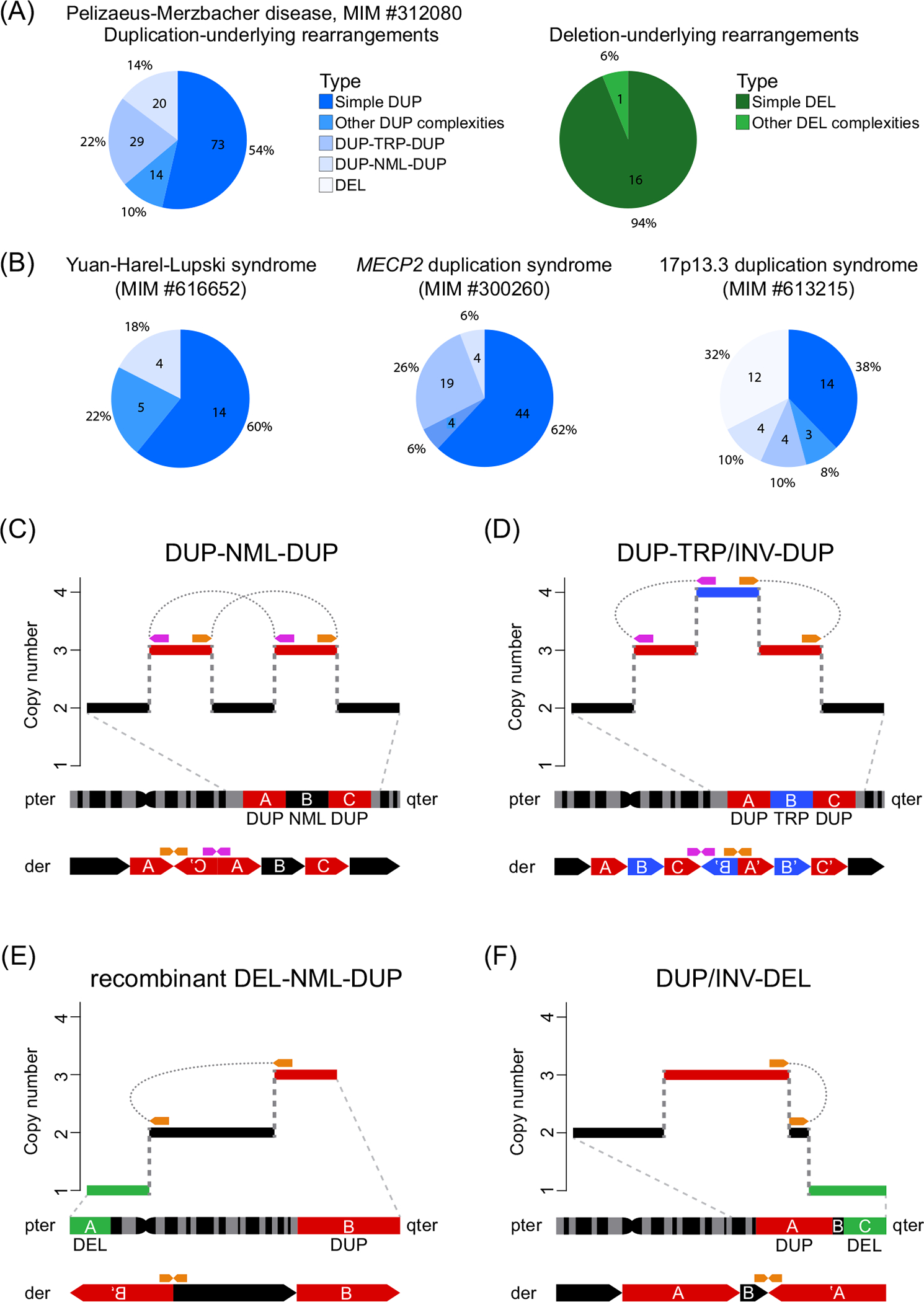
(A) Pie charts showing the proportion and absolute numbers of duplications (left) [50,52,92–95] and deletions (right) [96] causing Pelizaeus–Merzbacher disease. (B) Deletions and duplications in complex genomic events causing Yuan–Harel–Lupski syndrome [97], MECP2 duplication syndrome [41,51,98], and 17p13.3 duplication syndrome [49]. (C) Copy number signature of DUP–NML–DUP (interspersed duplications). One of four predicted DUP–NML–DUP genomic structures [49] is displayed at the bottom; this specific type was experimentally observed in pericentric inversions [10,99]. (D) Copy number signature of the DUP–TRP/INV–DUP CGR [inverted triplication (blue) flanked by duplications (red)]. The derivative structure displayed in the bottom was proposed from aCGH, Sanger sequencing, and FISH experiments. It is the first structure identified in probands affected with MECP2 duplication syndrome or Pelizaeus–Merzbacher disease [41]. Recently, three alternative structures were proposed based on experimental observations in cancer genomes [55]. (E) Copy number signature of a recombinant DEL–NML–DUP (telomeric deletion followed by a copy number neutral chromosome with a telomeric duplication). The duplicated sequence (red) is inverted and inserted at the location of the deletion (green). This rearrangement results from a meiotic recombination in a parent carrying a heterozygous copy number neutral pericentric INV [10] which will be resolved as a recombinant chromosome with a DEL–NML–DUP structure. (F) Copy number signature of DUP/INV–DEL (inverted duplication adjacent to terminal deletion). This structure results from chromosomes with terminal deletions further repaired by a fold-back mechanism mediated by short segments of homology creating a spacer [B] between the inverted duplications [A]. This type of structure can also be resolved as a translocation (inverted duplication translocation) or as a ring chromosome, both of which can be generated through a breakage–fusion–bridge cycle [100]. Purple and orange arrows represent the location of junctions in the reference genome. Abbreviations: aCGH, array comparative genomic hybridization; der, derivative chromosome; DUP, duplication; FISH, fluorescence-in-situ-hybridization; INV, inversion; MIM, Mendelian Inheritance in Man; NML, normal; pter, end of the short arm (p) of the chromosome; qter, end of the long arm (q) of the chromosome; TRP, triplication.
