Figure 6.
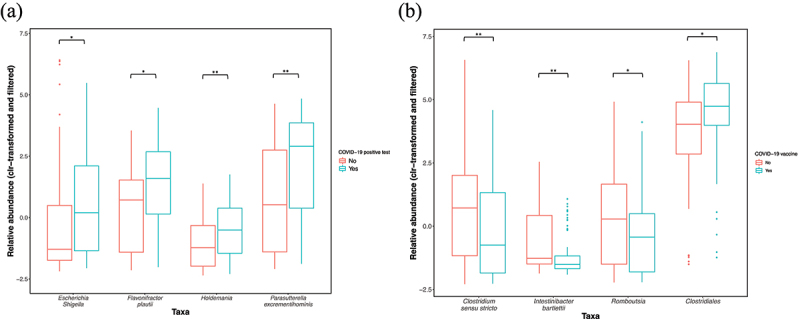
(a) Associations between a previous COVID-19 infection and the relative abundance of Escherichia-Shigella (mdn = 0.15 versus mdn = −1.36) (GLM, p = 0.004, r = 0.23, n = 198), Parasutterella excrementihominis (mdn = 2.85 versus mdn = 0.48) (GLM, p = 0.0003, r = 0.25, n = 198), Flavonifractor plautii (mdn = 1.52 versus mdn = 0.43) (GLM, p = 0.002, r = 0.21, n = 198) and Holdemania (mdn = −0.60 versus mdn = −1.29) (GLM, p = 0.0003, r = 0.24, n = 198). (b.) Associations between COVID-19 vaccine administration and the relative abundances of Clostridium sensu stricto (mdn = −0.79 versus mdn = 0.65) (GLM, p = 0.005, r = 0.22, n = 198), Intestinibacter bartlettii (mdn = −1.58 versus mdn = −1.33) (GLM, p ≤ 0.002, r = 0.3, n = 198), Romboutsia (mdn = −0.48 versus mdn = 0.22) (GLM, p = 0.01, r = 0.22, n = 198) and the Clostridiales order (mdn = 4.61 versus mdn = 3.77) (GLM, p = 0.01, r = 0.25, n = 198). Y-axes show the clr-transformed relative abundances of the taxa. The solid line indicates the median, lower and upper bounds of boxes indicate the first and third quartiles, respectively; whiskers indicate the 1.5 IQR beyond the upper and lower quartiles. Dots represent outlier data points. Sample sizes: previous COVID-19 infection YES n = 42, previous COVID-19 infection NO n = 156. COVID-19 vaccine administered YES n = 90, COVID-19 vaccine administered NO n = 108. Significance * for p ≤ 0.05, ** for p ≤ 0.005.
