Figure 4.
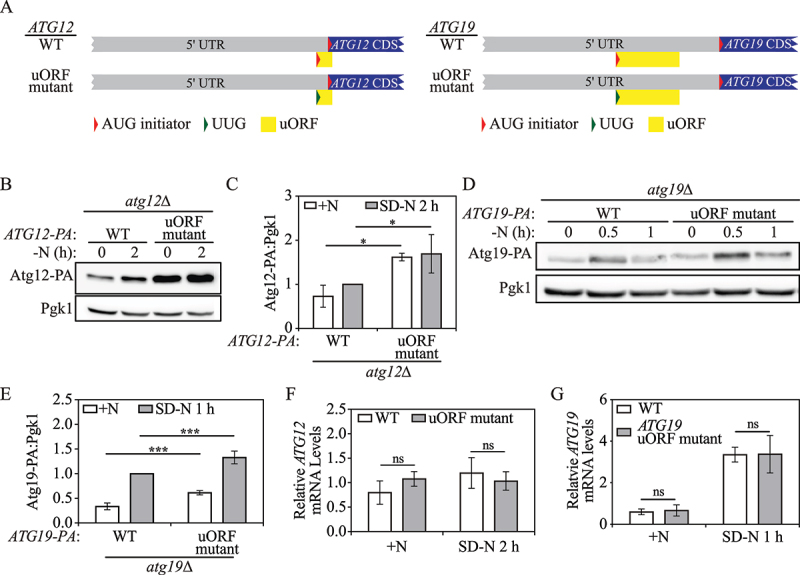
Yeast Atg12 and Atg19 translation are mediated by a single inhibitory uORF in the corresponding 5’ UTR. (A) Schematic representation of partial ATG12 or ATG19 mRNA transcripts with a potential functional uORF identified by ribosome profiling (yellow box). The ATG12 uORF has a start codon (AUG) in the 5’ UTR, and an in-frame stop codon preceding the end of the ATG12 CDS. The ATG19 uORF has a start codon (AUG) in the 5’ UTR, and a stop codon preceding the ATG19 CDS. The mutations made in the start codon of the ATG12 or ATG19 uORF in the plasmid constructs are shown as indicated (represented as a change from a red triangle to a green triangle). (B) Atg12-PA levels were measured by western blot in WT and ATG12 uORF mutant cells under growing conditions and after 2 h of nitrogen starvation. Pgk1 was used as a loading control. (C) The ratio of Atg12-PA to Pgk1 was quantified. Mean±SD of n = 3 independent experiments are shown as indicated. ANOVA; *: p < 0.05. (D) Atg19-PA levels were measured by western blot in WT and ATG19 uORF mutant cells under growing conditions and after 0.5 h and 1 h of nitrogen starvation. (E) The ratio of total Atg19-PA to Pgk1 under growing and 1-h nitrogen-starvation conditions was quantified. Mean±SD of n = 4 independent experiments are shown as indicated. ANOVA; ***: p < 0.005. (F) ATG12-PA mRNA levels were determined in WT and ATG12 uORF mutant cells under growing and 2-h nitrogen-starvation conditions by RT-qPCR. Mean±SD of n = 3 independent experiments are shown as indicated. ANOVA; ns: not significant. (G) ATG19-PA mRNA levels were determined in WT and ATG19 uORF mutant cells under growing and 1-h nitrogen-starvation conditions by RT-qPCR. Mean±SD of n = 3 independent experiments are shown as indicated. ANOVA; ns: not significant. SD-N, synthetic minimal medium lacking nitrogen; +N (YPD), yeast extract-peptone-dextrose; WT, wild type; PA, protein A.
