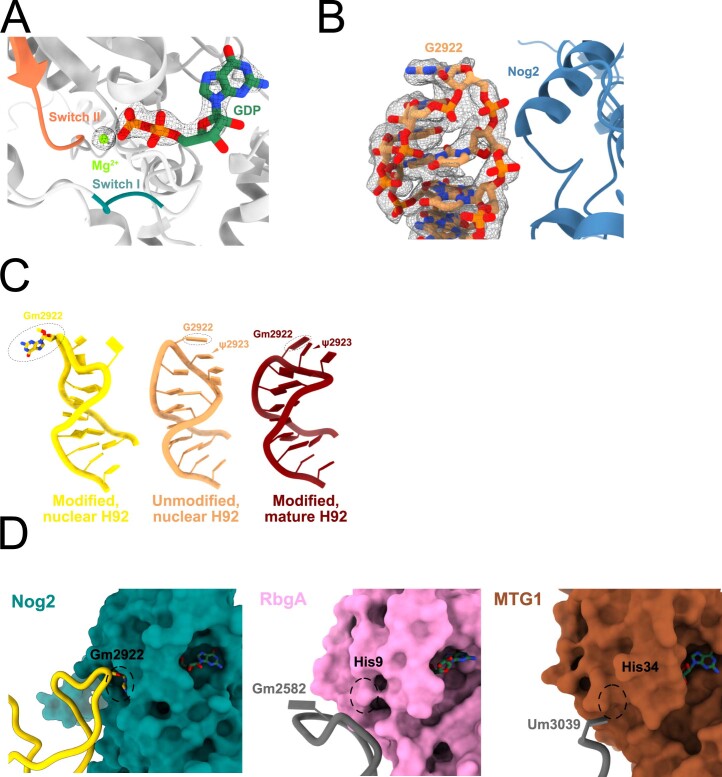
Extended Data Fig. 7. Details of the unmodified G2922 structure and structural comparison between Nog2 and orthologs.
A. Active site of Nog2 in complex with pre-60S lacking G2922 methylation. Densities for GDP, Mg2+ and ordered regions Switch I and Switch II are clearly observed. B. Cryo-EM map corresponding to H92 in the unmodified G2922 structure. C. Cartoon comparisons between H92 with a modified Gm2922 engaged with Nog2 (left, accession no. PDB 3JCT), an unmodified G2922 (middle, this work) and the mature H92 from the yeast ribosome crystal structure (right, accession no. PDB 4V88). D. Comparison between the active sites of Nog2 (left, accession no. PDB 3JCT), bacterial ribosome biogenesis GTPase RbgA (middle, accession no. PDB 6PPK) and human mitoribosome biogenesis GTPase MTG1 (right, accession no. PDB 7PD3). The position of Gm2922, or locations of histidine residues implicated in catalysis, are indicated by dashed circles.