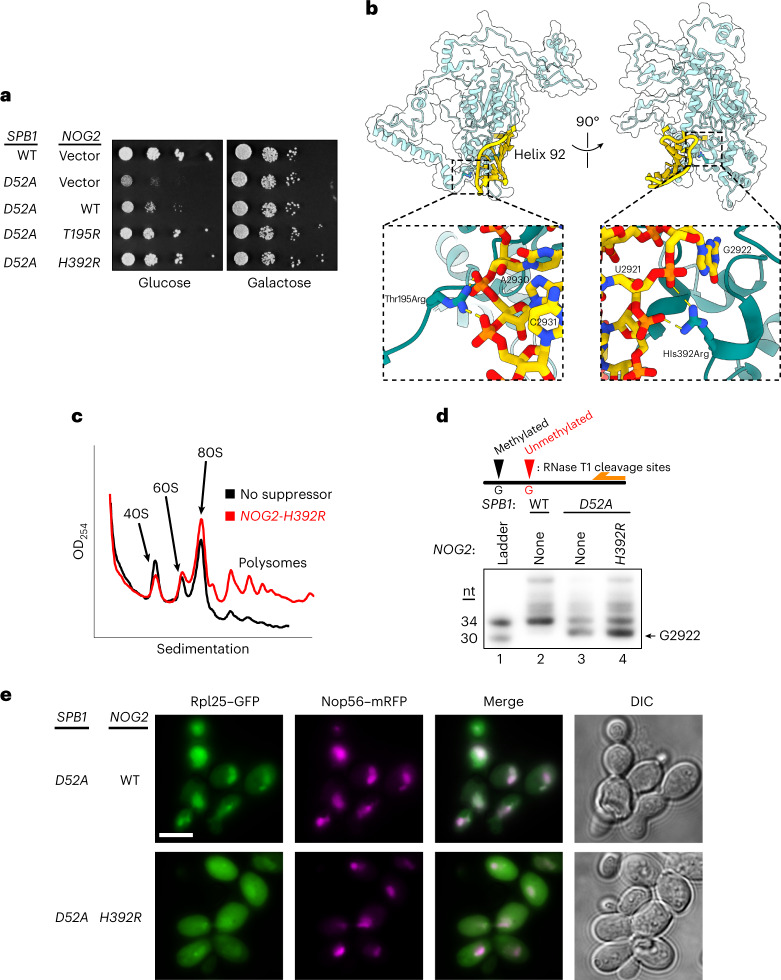
Fig. 3. Single amino acid changes in Nog2 restore robust growth and nuclear export in the absence of Gm2922.
a, Genetic suppression of a lack of Gm2922 by the indicated NOG2 alleles in a repressible SPB1 strain (glucose, endogenous SPB1 repressed; galactose, expressed). b, Cartoon and surface representation of Nog2, with suppressing amino acid changes modeled (accession no. PDB 3JCT). Left inset, suppressing amino acid change Thr195Arg modeled to show new electrostatic interactions with the phosphates of the H92 bases A2930 and C2931. Right inset, suppressing amino acid change His392Arg modeled similarly to show new electrostatic interactions with the phosphates of the H92 bases U2921 and G2922. c, Sucrose gradient sedimentation profile from spb1-D52A cells suppressed by NOG2-H392R, contrasted with empty vector (black trace same as shown in Fig. 2c). d, Primer extension assay to probe 2′-O-methylation at G2922 in WT cells, cells expressing methyltransferase-defective spb1-D52A and spb1-D52A suppressed by NOG2-H392R. Lanes 2 and 3 are the same samples as loaded in Fig. 2d. The experiment was repeated twice with similar results. e, Fluorescence microscopy to monitor location of large subunit (Rpl25–eGFP) and nucleolus (Nop56–mRFP) when spb1-D52A is suppressed by WT NOG2 or NOG2-H392R. The experiment was repeated twice with similar results. Scale bar, 5 µm.