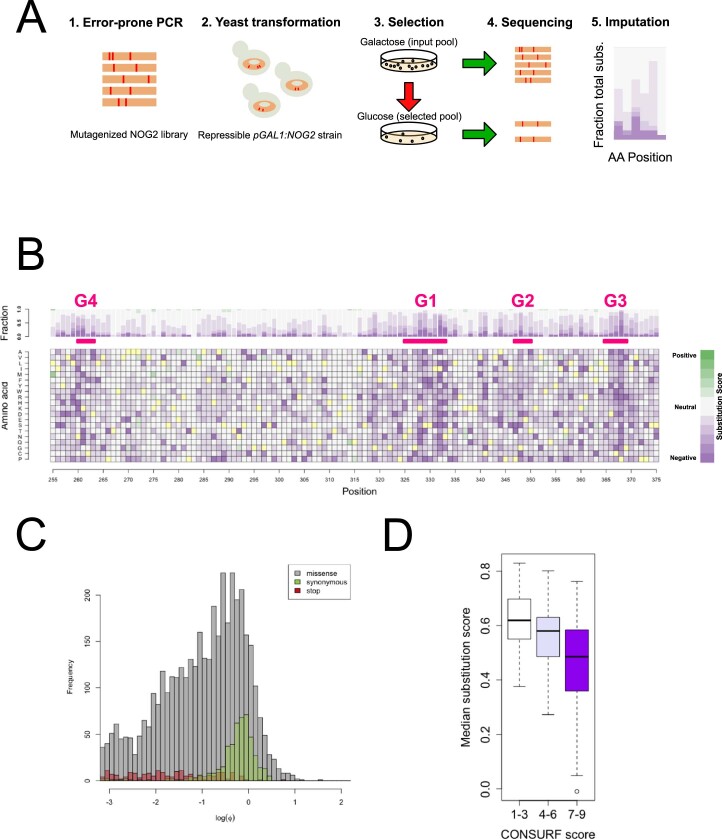
Extended Data Fig. 1. Massively parallel mutagenesis of NOG2.
A. Scheme for high throughput mutagenesis and analysis of functional variants of NOG2. B. Complete fitness landscape for variants in the core GTP-binding motifs of NOG2 (G4, G1, G2, G3; amino acids 260-369). Yellow squares indicate the WT amino acid identity. C. Distributions of observed fitness scores (Φ) for nonsense (red), missense (gray) and synonymous (green) mutations. D. Correlation between per-residue median fitness score and ConSURF 41 score (evolutionary conservation: scores 1-3, low; scores 4-6, medium; scores 7-9, high conservation). The statistical analysis used was performed in TileSeq and described in Weile et al. (ref. 8).