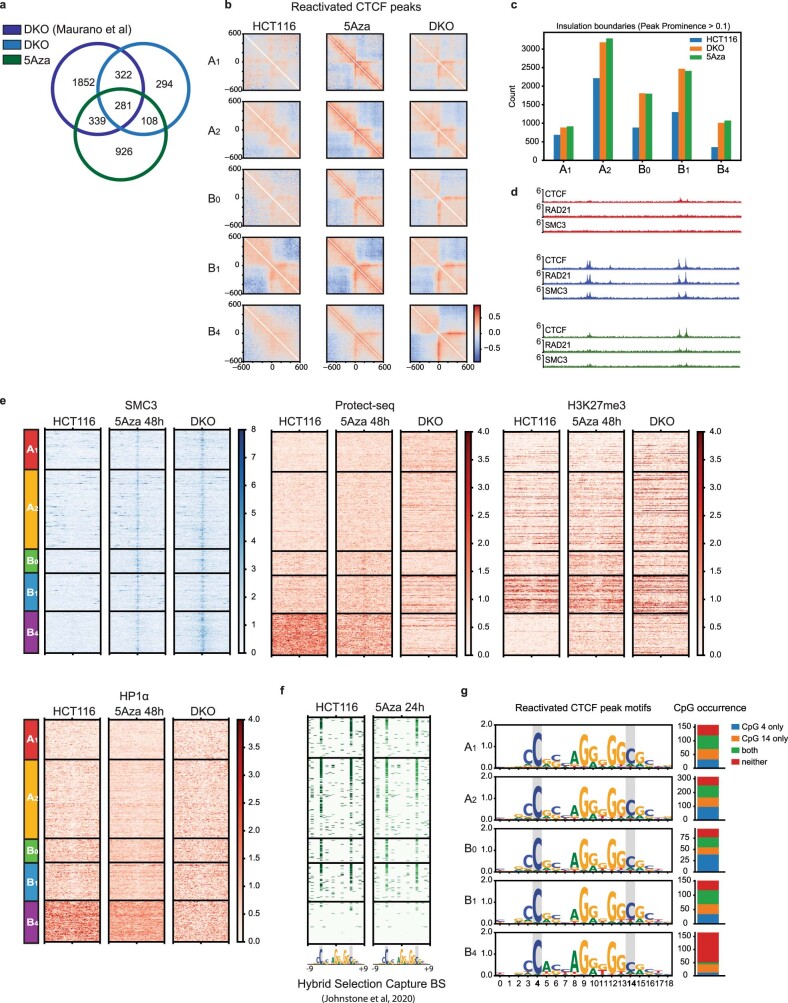
Extended Data Fig. 10. Reactivated CTCF sites.
(a) Venn diagram of CTCF peaks in DKO (this study), 5Aza (this study), and DKO (Maurano et al.61). Union between CTCF peaks used to define reactivated CTCF sites. (b) Average observed/expected Hi-C maps around reactivated CTCF binding sites within each IPG centered at CTCF motifs oriented as indicated in HCT116 (left), 5Aza (center), and DKO (right) cells. (c) Quantification of total number of insulating loci with peak prominence score > 0.1 per IPG. (d) Example region (chr11:39–40 Mb) of reactivated CTCF sites blocking cohesin (RAD21 and SMC3). (e) Stacked heatmaps of reactivated CTCF sites for HCT116, 5Aza, and DKO cells centered on the CTCF motif displaying ChIP-seq signal for SMC3 (upper left), Protect-seq (middle), H3K27me3 (right), and HP1α (lower left) flanked by ±5 kb and segregated by IPG. (f) Similar to Fig. 7c. Stacked heatmaps around reactivated CTCF site core motifs (19 bp) for HCT116 and 5Aza-treated cells displaying fraction CpG methylation using hybrid selection capture bisulfite sequencing data from (Johnstone et al.77). (g) Left: sequence logos for the reactivated CTCF motifs in each IPG. Right: frequencies of CpG occurrence at motif positions 4 and 14 in each set of reactivated CTCF sites. Note: nucleotides 4 and 14 depend on the motif start, other publications refer to these CpG nucleotides as 2 and 12 (for example Hashimoto et al.63) or 1 and 11 (for example Wang et al.33).