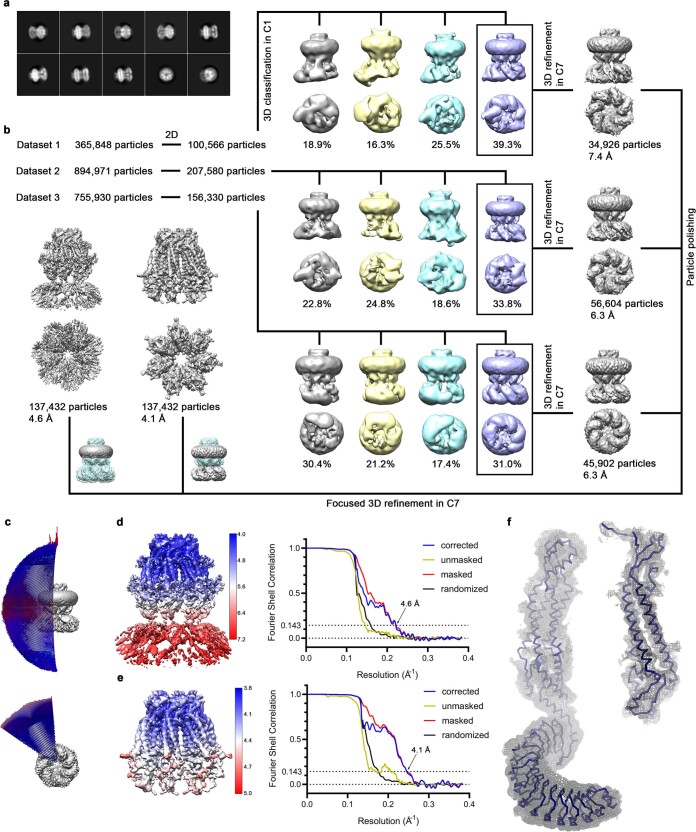
Extended Data Fig. 3. Cryo-EM structure of LRRC8C.
a, Representative 2D class averages of the LRRC8C channel. b, Data processing workflow. Particles from three datasets were cleaned-up during 2D classification and selected class averages showing high resolution features were used in the subsequent 3D classification with no symmetry applied. In each dataset, one out of four classes showed a symmetric pore domain and seven, albeit flexible, LRRDs. The distribution of particles (%) is indicated. Particles assigned to the boxed classes were used in independent refinements with C7 symmetry applied. As all three reconstructions show the same features, all particles belonging to these refined models were pooled and subjected to per-particle motion correction. Polished particles were subsequently used as input for C7-symmetrized focused refinement of the full-length channel and TM region. Insets show the masked regions during refinement. c, Angular distribution plot of all particles included in the final reconstruction of the full-length complex. The length and color of cylinders correspond to the number of particles with respective Euler angles. d, e, Final 3D reconstruction colored according to local resolution (left) and FSC plot (right) of the final refined unmasked (yellow), masked (red), phase-randomized (black) and corrected for mask convolution effects (blue) cryo-EM density map of the LRRC8C channel. The resolution at which the FSC curve drops below the 0.143 threshold is indicated. d, Full-length complex and e, masked PD. f, Cα representation of a single subunit of LRRC8C with cryo-EM density of the entire protein at 4.6 Å (contoured at 5 σ, left) and of its PD at 4.1 Å (contoured at 10 σ, right) shown superimposed as gray mesh.