Extended Data Fig. 2 |. ZNF579 targets significantly overlap with the differentially expressed genes in SARS-CoV-2 infected patient samples.
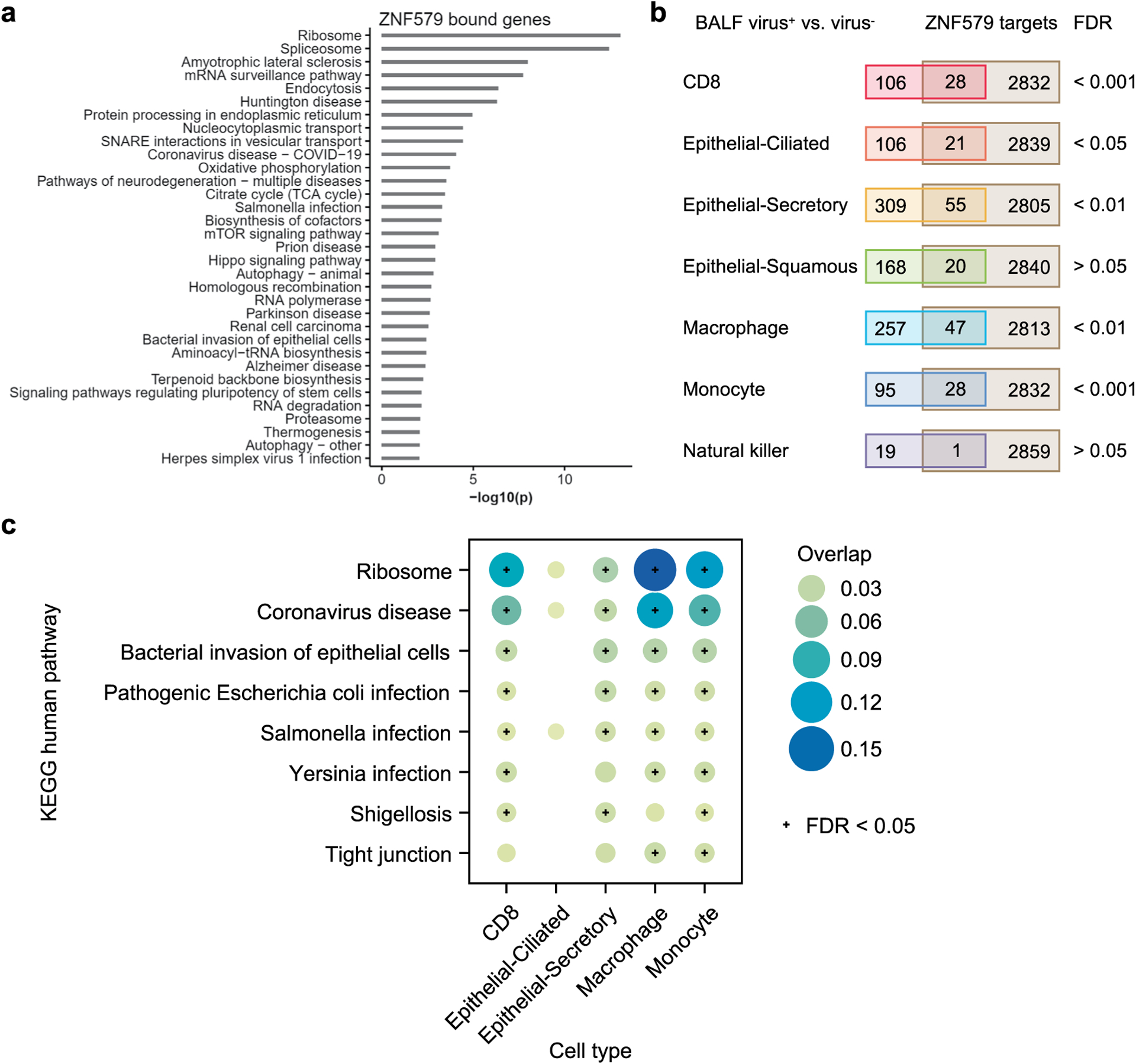
(a) Enriched KEGG pathways of genes associated with ZNF579 binding by ChIP-seq (ENCODE:ENCSR018MQH). Genes were considered to be bound by ZNF579 if a ChIP-seq peak overlapped with the promoter region (-1000 to transcription start site). (b) Overlap of ZNF579 targets and differentially expressed genes (DEGs) in bronchoalveolar lavage fluid (BALF) SARS-CoV-2+ vs. SARS-CoV-2− samples. See Methods for the source of the single-cell dataset. Fisher’s exact tests show that the overlaps are significant (FDR < 0.05) for five cell types, including CD8, epithelial-ciliated, epithelial-secretory, macrophage and monocyte. (c) The enriched pathways of the overlapping ZNF579 targets and DEGs in the five cell types. Pathways that are significantly enriched in at least two cell types are shown.
