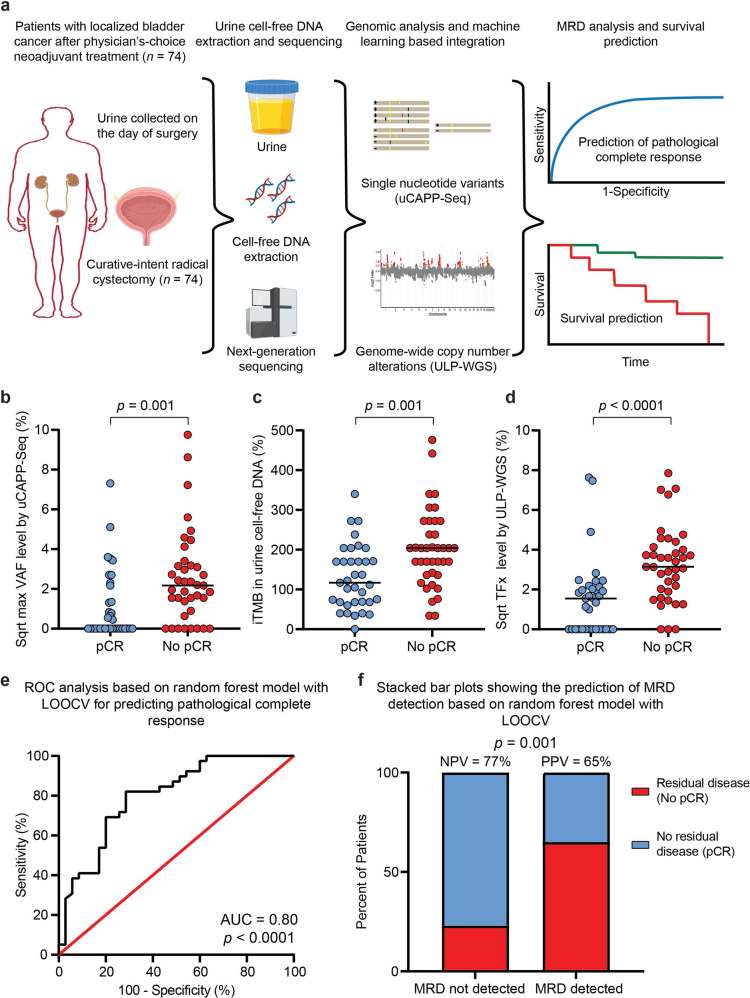
Fig. 1. Pathologic complete response prediction using a random forest model based on urine tumor DNA.
a Urine was collected prospectively from 74 localized bladder cancer patients pre-operatively on the day of curative-intent radical cystectomy after physician’s-choice neoadjuvant treatment. Urine cell-free DNA was sequenced by uCAPP-Seq (for single nucleotide variants) and ULP-WGS (for genome-wide copy number alterations) and then correlated with residual tumor in the surgical resection specimen and with patient survival. This figure panel was created with BioRender.com. b SNV-derived maximum VAFs, c inferred tumor mutational burden, and d CNA-derived tumor fraction levels in urine cell-free DNA from patients with localized bladder cancer. Scatter plots display these three different urine cell-free DNA metrics, stratified by pathologic complete response status, with significance determined by the Mann–Whitney U-test. VAF and CNA-derived tumor fraction data are shown after square root transformation. e ROC analysis of random forest model integrating urine tumor DNA metrics and other pretreatment clinical variables (Supplementary Fig. 5). ROC curve demonstrating the model’s performance for predicting pCR after LOOCV (AUC = 0.80, p < 0.0001). f Stacked bar plot depicting NPV and PPV of the random forest model with LOOCV, with significance determined by the Fisher’s exact test. AUC area under the curve, cfDNA cell-free DNA, CNA copy number alteration, iTMB inferred tumor mutational burden, LOOCV leave-one-out cross-validation, max maximum, MRD molecular residual disease, NPV negative predictive value, pCR pathologic complete response, PPV positive predictive value, ROC receiver operating characteristic, SNV single nucleotide variant, Sqrt square root, TFx tumor fraction, uCAPP-Seq urine cancer personalized profiling by deep sequencing, ULP-WGS ultra-low-pass whole genome sequencing, VAF variant allele frequency.