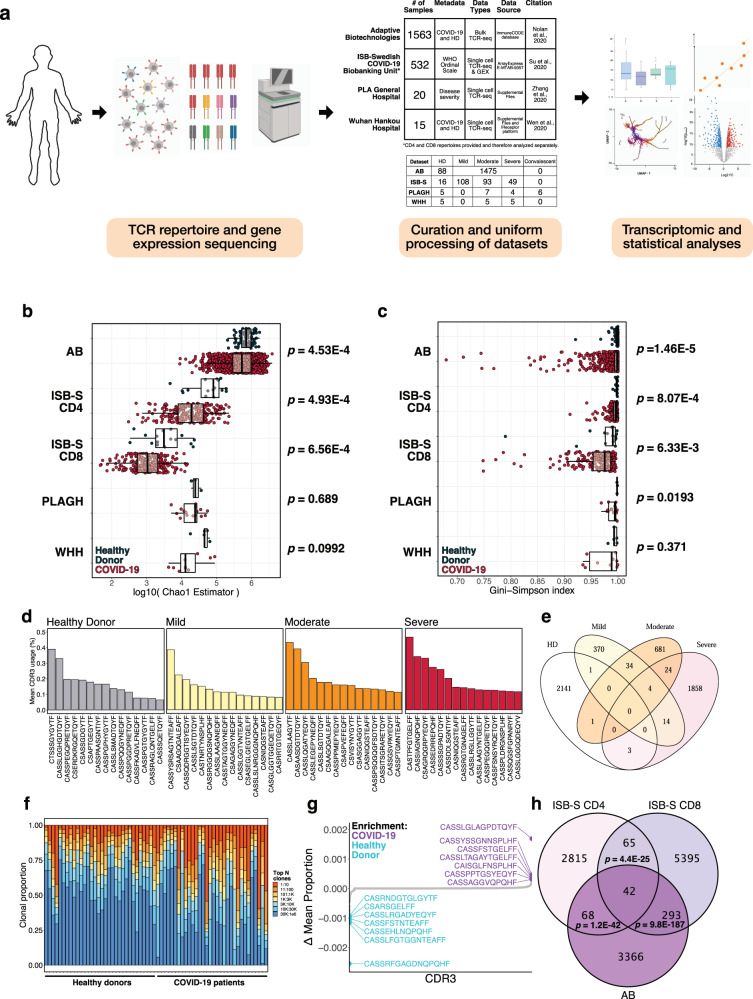
Fig. 1. Analysis of TCR repertoires from COVID-19 patients and healthy donors reveal trends in CDR3 gene usage and diversity.
a Schematic detailing curation and analysis of TCR repertoire datasets from healthy donors and COVID-19 patients. Sequencing data was obtained from Adaptive Biotechnologies (AB, n = 1574), ISB-Swedish COVID-19 Biobanking Unit (ISB-S, n = 266, CD4 and CD8 repertoires), PLA General Hospital (PLAGH, n = 20), and Wuhan Hankou Hospital (WHH, n = 15). Created with BioRender.com. b Boxplot of Chao1 indices for COVID-19 patients and healthy donors for each repertoire dataset. P-values were obtained using the two-sided Wilcoxon rank sum test. c Boxplot of Gini-Simpson indices for COVID-19 patients and healthy donors for each repertoire dataset. P-values were obtained using the two-sided Wilcoxon rank sum test. d Bar plots showing the top 15 mean CDR3 usages for patients in the ISB-S CD4 dataset grouped by disease severity (healthy donor = 16, mild = 108, moderate = 93, severe = 49). e Venn diagram showing overlap of top mean CDR3 usages (proportion threshold = 0.0001) for patients in the ISB-S CD4 dataset grouped by disease severity. The CDR3 sequences enriched in the COVID patients have overlap among mild, moderate, and severe patients, while minimal overlap is observed between healthy donors and COVID-19 patients. f Bar plot depicting relative abundance for groups of top clonotypes for a random sample of repertoires (healthy donors = 32, COVID-19 = 32) from AB dataset, with relative overrepresentation of specific clonotypes in COVID-19 patients. g Dotted waterfall plot of CDR3 gene usage differentials between COVID-19 patients and healthy donors (delta mean proportion) in AB dataset. Purple dots are CDR3 sequences enriched in COVID-19; light blue dots are CDR3 sequences enriched in healthy donors; gray dots are all other CDR3 sequences. h Venn diagram showing overlap of COVID-19 enriched CDR3 sequences for patients in the ISB-S CD4, ISB-S CD8, and AB datasets (thresholds 0.0001 for ISB-S samples, 0.00001 for AB samples). P-values for overlap significance calculated using hypergeometric test.