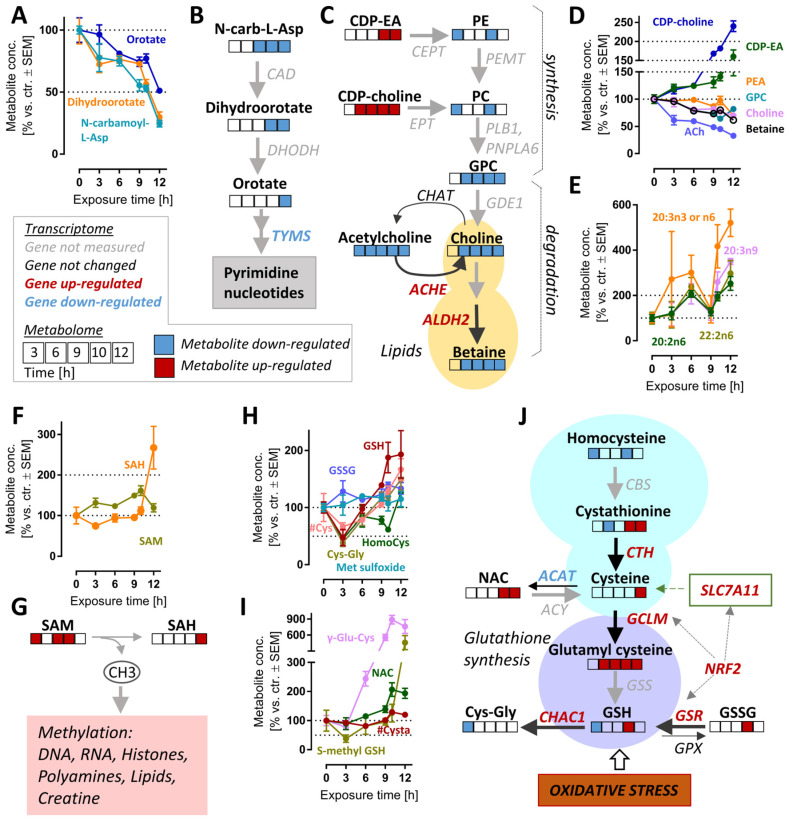
Figure 6.
Cell responses relevant to late time points after MG-132 treatment of dopaminergic neurons. Metabolome and transcriptome samples were obtained as detailed in Figure 1. (A) Time-dependent changes of pyrimidine de novo synthesis intermediates and (B) overview of this pathway with mapped omics responses. (C) Choline metabolism pathway diagram and time-dependent changes (D) also incorporating the transcriptomic changes of genes coding for relevant enzymes (9 h). (E) Levels of polyunsaturated fatty acids (PUFA) over time. (F,G) Time-dependent changes of the methyl donor S-adenosyl methionine (SAM) and its metabolite S-adenosyl homocysteine (SAH). (H,I) Time-dependent changes of glutathione-related pathways. (J) Graphical overview of GSH metabolism, with mapped omics responses. A complete set of data for these pathways is provided in Figure S8. A full overview of captured metabolites is found in Table S1. A full overview of targeted transcripts is provided in Table S2. Metabolite data are means ± SEM of independent replicates. For MG-132 treatments, 3 different samples were prepared. For controls (DMSO), 4 replicates were prepared to provide for more robust baseline data. #, metabolite quantified by amino acid analyzer; ACAT, acetyl-CoA acetyltransferases; ACh, acetylcholine; ACHE, acetylcholinesterase; ACY, aminoacylase; ALDH2, aldehyde dehydrogenase 2 family member; CAD, carbamoyl-phosphate synthetase 2, aspartate transcarbamylase and dihydroorotase; CBS, cystathionine beta-synthase; CDP-choline, cytidine diphosphate-choline; CDP-EA, cytidine diphosphate-ethanolamine; CEPT, choline/ethanolamine phosphotransferases; CHAT, choline O-acetyltransferase; CTH, cystathionine gamma-lyase; Cys, cysteine; Cysta, cystathionine; DHODH, dihydroorotate dehydrogenase; EPT, CDP-ethanolamine:diacylglycerol ethanolaminephosphotransferase; GCLM, glutamate-cysteine ligase modifier subunit; GDE1, glycerophosphodiester phosphodiesterase 1; GPC, glycerylphosphorylcholine; GPX, glutathione peroxidases; GSH, oxidized glutathione; GSR, glutathione-disulfide reductase; GSS, glutathione synthetase; GSSG, reduced glutathione; HomoCys, homocysteine; Met sulfoxide, methionine sulfoxide; NAC, N-acetyl cysteine; N-carb-L-Asp, N-carbamoyl-L-aspartate; NRF2, NFE2-like BZIP transcription factor 2; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PEMT, phosphatidylethanolamine N-methyltransferase; PLB1, phospholipase B1; PNPLA6, patatin-like phospholipase domain containing 6; SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine; SLC7A11, cystine/glutamate transporter; TYMS, thymidylate synthetase.