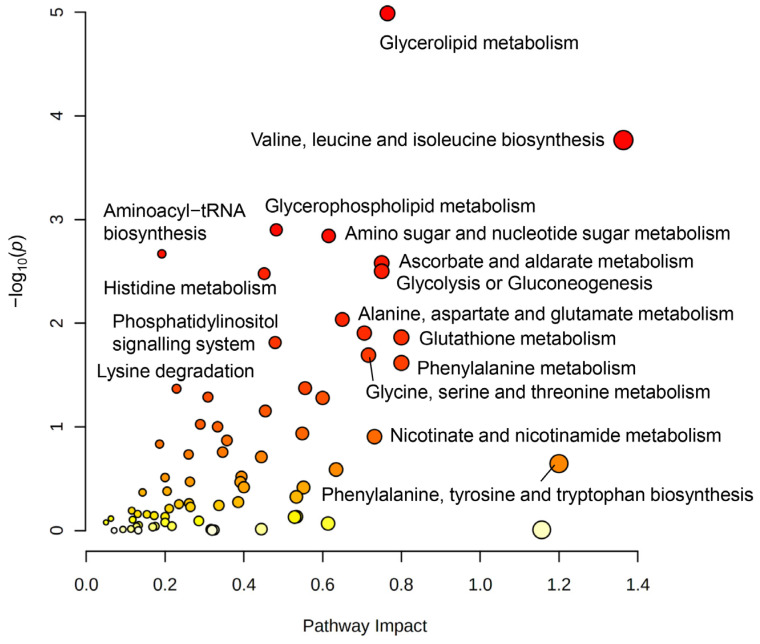
Figure 6.
Joint pathway analysis of 1H-NMR spectroscopy-based metabolomics and RNA-seq gene fold change data. Pathway analysis indicates the most significantly changed metabolic pathways (the node color scheme is based on p-values from yellow dots = low significance to red dots = high significance and the node radius is determined based on the pathway impact value). Glycerolipid metabolism, branched-chain amino acid (BCAA) metabolism of valine, leucine, and isoleucine biosynthesis, glycerophospholipid metabolism, amino sugar and nucleotide sugar metabolism, ascorbate metabolism, glycolysis or gluconeogenesis, and alanine, aspartate, and glutamate metabolism, were most affected based on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database. For joint-pathway analysis, data from metabolomics (n = 4) and RNA-seq (n = 4) were used.