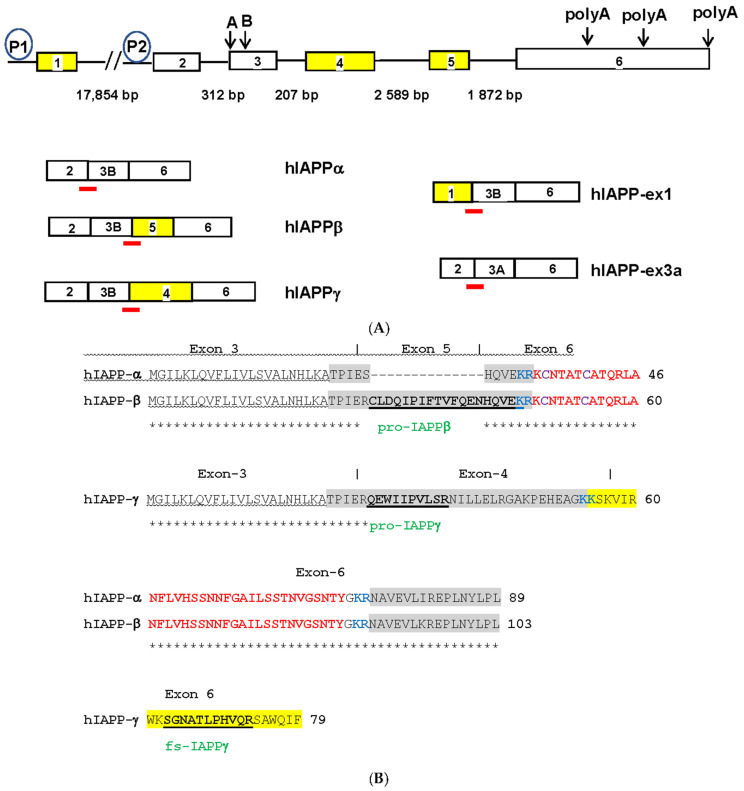
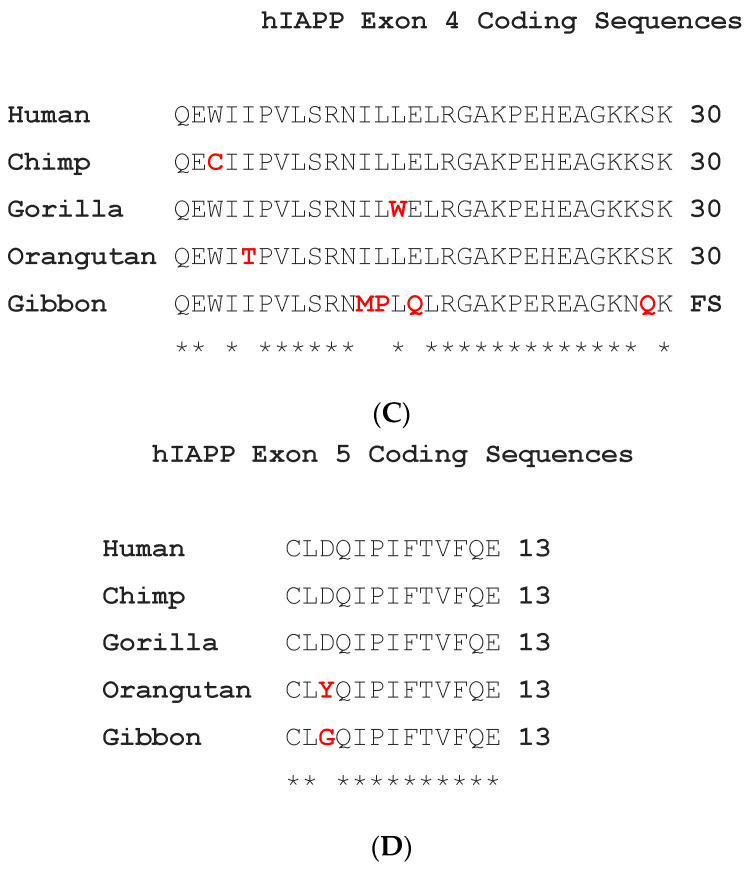
Figure 1.
(A) Human IAPP gene structures and alternatively spliced isoforms. Open boxes represent exons, solid lines represent introns of different sizes in base pairs (bp) and P1 and P2 in circles are alternative promoters. Yellow boxes represent hominid-specific IAPP exons. Downward arrows and capital letters are at the intra-exonal splicing donor sites in exon 3 and alternative polyadenylation sites in exon 6. The red bars are exon junctional TaqMan probes that hybridize specifically to various IAPP isoforms. The spliced transcripts of hIAPP isoforms are named and below the gene structure. hIAPP-ex1 and -ex3a are not expressed and are expressed at low levels in islets, respectively. (B) Alignment of hIAPPα, hIAPPβ, and hIAPPγ amino acid sequences. Homologous amino acids are marked by asterisks, signal peptide underlined by wavy lines, prohormone regions by gray highlight, the convertase dibasic amino acids by blue lettering, mature IAPP37 by red lettering, bisulfide bond cysteine (C) by purple lettering, IAPP25 by yellow highlight, the proteolytic peptide names by green lettering, and the sequences are bolded and underlined. Exon numbers are above the sequences and demarcated by the | sign. (C) Alignment of exon 4 of the hIAPPγ CDS. (D) Alignment of exon 5 of the hIAPPβ CDS. The amino acids coded by nonsynonymous substitutions are shown in red lettering.