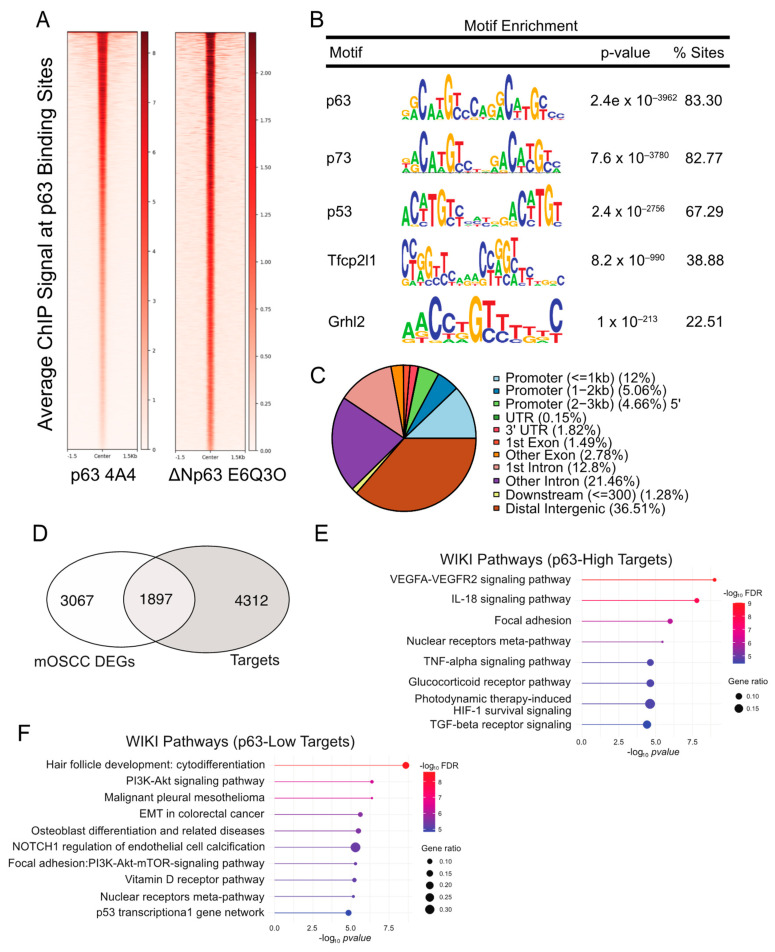
Figure 2.
Identification of the p63 global targets in B7E11 cells by ChIP-seq. (A) Heat map of the average ChIP-seq signals from p63 binding sites for two p63 antibodies across the genome. (B) Top transcription factor motifs derived from CentriMo motif analysis on B7E11 consensus p63 ChIP peaks. (C) Distribution pattern of genomic features associated with p63 binding sites across the genome. (D) Integration of the identified p63 cistrome with the previously identified cell line-based DEG dataset. Enriched WikiPathways identified for p63high (E) and p63low (F) target genes.