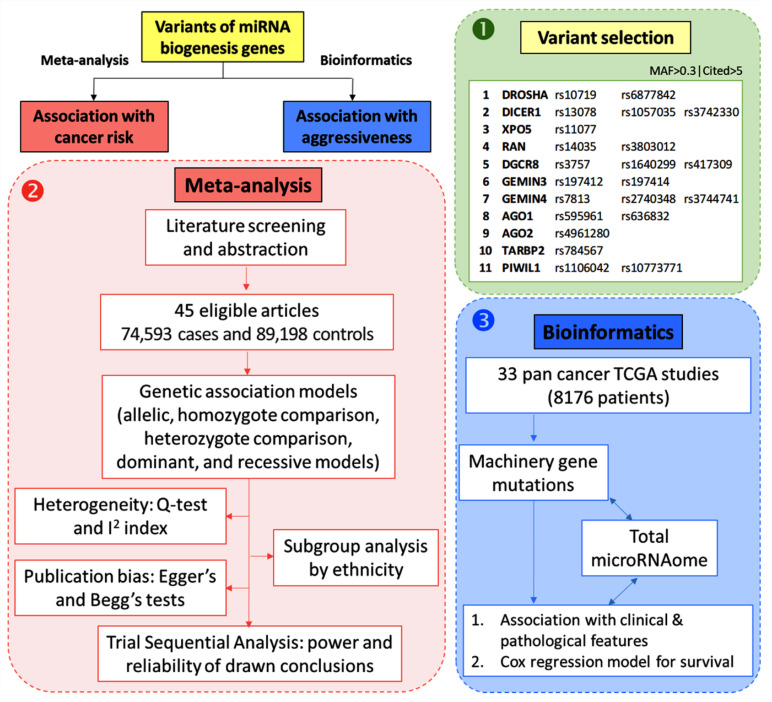
Figure 2.
Workflow followed for in silico data analysis and meta-analysis. We followed three steps: (1) Selection of genes enrolled in microRNA machinery canonical pathway followed by a selection of genetic variants with five or more published articles and minor allele frequency (MAF) of more than 0.3 after identification of 11 genes, including 22 single nucleotide polymorphisms. (2) Meta-analysis was performed to pool the results of 45 published articles. Pairwise comparisons were carried out for each genetic association model to identify the association between gene polymorphism and cancer risk. Next, heterogeneity analysis, subgroup analysis, trials sequential analysis, and publication bias were assessed. (3) To explore the association between genetic variants and cancer prognosis, transcriptomic and genomic data of 33 different types of cancer were retrieved from the TCGA database. Genetic mutations of the 11 studied genes were screened in association with the expression of corresponding genes and the total microRNAs expression in each patient. The Mann–Whitney U and Kruskal–Wallis tests were employed to test the association between gene mutation and clinicopathological features, including lymph node metastasis, distant metastasis, and pathological grade. Survival analysis was performed using Cox regression analysis and Kaplan–Meier curve plots were generated. Both univariate and multivariate analyses using total miRNAome level were conducted.