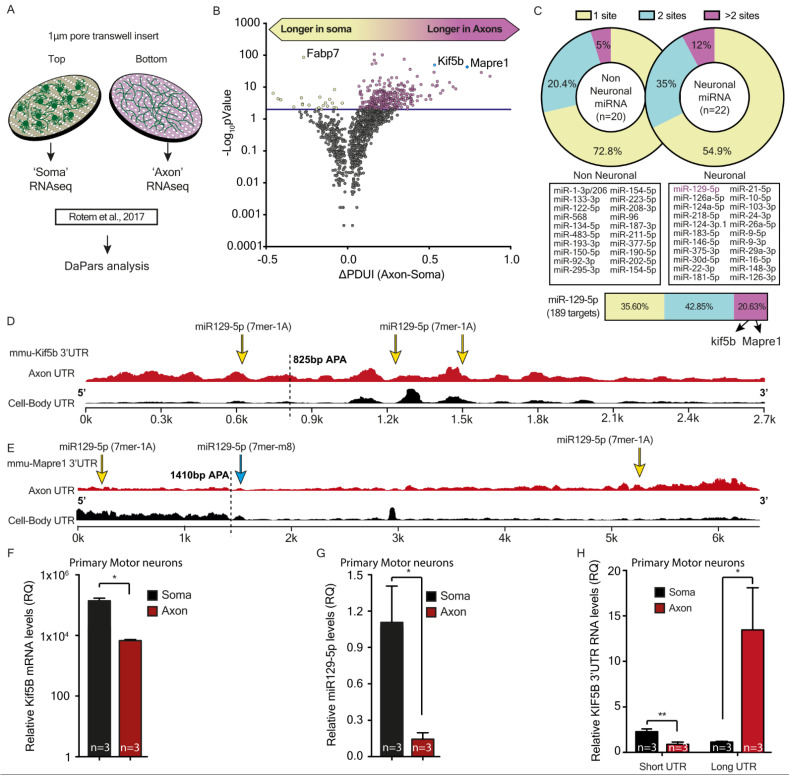
Figure 4.
Kif5b mRNA transcript variant length and expression at axon versus cell-body motor neurons. (A) Illustration of the transwell insert method used for isolation of axonal RNA from primary motor neuron (MNs). (B) A volcano plot demonstrating results of the Dynamitic analysis of Alternative Poly-Adenylation from RNA-seq (DaPars) analysis for the ΔPDUI, representing the differences in the short vs. long 3′UTR ratios in axons vs. cell bodies. Significantly elongated 477 mRNA transcripts are colored in red. Kif5b and Mapre1 (blue) were chosen for further validations. (C) Upper panel: analysis of the percent of long 3′UTR transcripts containing 1, 2, or >2 target sequences for neuronal versus non-neuronal miRNAs. The middle panel shows the list of neuronal and non-neuronal miRNAs tested. The lower panel details the specific percent of miR-129-5p targets within the long 3′UTR transcripts that contain 1, 2, or >2 target sequences. Two transcripts with more than two target sequences for miR-129-5p are highlighted: Kif5b and Mapre1. (D) The diagram compares 3′UTR coverage of the Kif5b transcript in axons (red) versus cell-bodies (black). The alternative polyadenylation (APA) site is marked with a black dashed line. The yellow arrows indicate positions of miR-129-5p target sequences. (E) The diagram compares 3′UTR coverage of Mapre1 transcript in axons (red) versus cell-bodies (black). The APA site is marked with a black dashed line. The yellow arrows indicate positions of miR-129-5p target sequences. (F–H) RT-qPCR analysis of (F) Kif5b coding sequence, (G) mature miR-129-5p, or (H) shortened and elongated Kif5b 3′UTR expression in axons and cell bodies of primary MNs. Normalized fold change (FC) is shown in Kif5b and miR-129-5p expression to GAPDH and miR-124a-3p, respectively. The 3′UTR expression in each compartment (H) is relative to the expression of the coding sequence exons. Unpaired students’ t-test. * p < 0.05, ** p < 0.01.