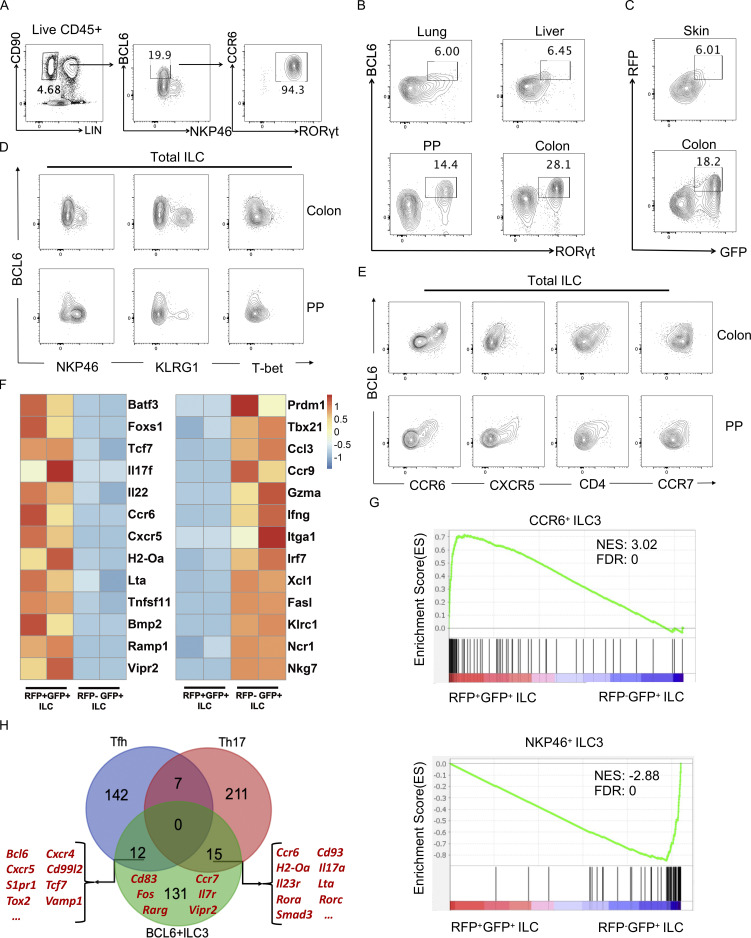
Figure 1.
BCL6 is expressed in ILC3 in multiple tissues. (A) Gating strategy for ILC in colon LPLs from B6 mice. Total ILC were gated on Lin−CD90+CD45+ live cells, in which a group of BCL6+NKP46− population was further gated and analyzed for CCR6 and RORγt expression by flow cytometry. (B) BCL6 and RORγt expression in total ILC isolated from B6 mice among indicated tissues at steady state. (C) BCL6 and RORγt expression in total ILC isolated from Bcl6RFP/RFPRorcGFP/+ mice among indicated tissues at steady state. (D) Co-staining of BCL6 versus NKP46, KLRG1, and T-bet in total ILCs isolated from the colon LP and PP of B6 mice. (E) Co-staining of BCL6 versus CCR6, CXCR5, CD4, and CCR7 in total ILCs isolated from the colon LP and PP of B6 mice. (F) Heat map of RNA-seq data for selected key signature genes of CCR6+ ILC3 (Schleussner et al., 2018; Talbot et al., 2020; Zhong et al., 2016; Zhong et al., 2018) and NKP46+ ILC3 (Gury-BenAri et al., 2016) in RFP+GFP+ vs. RFP−GFP+ ILCs obtained from the LPLs of Bcl6RFP/RFPRorcGFP/+ mice. The processed RNA-seq data was listed in Table S1, and raw data are available through GSE220438 on GEO DataSets. (G) GSEA analysis of literature signature genes of CCR6+ ILC3 (top panel) and NKP46+ ILC3 (bottom panel) enriched in RFP+GFP+ (BCL6+RORγt+) ILC versus RFP−GFP+ (BCL6−RORγt+) ILC gene sets. Gene sets were originated from RNA-seq data in F. See also Table S2 for detailed list of literature signature genes. NES, normalized enrichment score; FDR, false discovery rate. (H) Veen graph of top expressed signature genes in BCL6+ ILC3s (RNA-seq data of RFP+GFP+ ILC3 in F) and Tfh and Th17 cells (Liu et al., 2016; Tanaka et al., 2018). The listed signature genes were determined by overlapping top expressed genes from the three indicated gene pools and removing pseudogenes and housekeeping genes. Representative Tfh, Th17, and BCL6+ ILCs genes were highlighted in red (please also see Table S3 for detailed list of genes). Data are a representative of three (A, D, and E) or two (B and C) independent experiments.